name: tea-talk-feb25 class: title, middle ## Multi-fidelity active learning for scientific discoveries Alex Hernández-García (he/il/él) .center[ <a href="https://mila.quebec/"><img src="../assets/images/slides/logos/mila-beige.png" alt="Mila" style="height: 4em"></a> <a href="https://www.umontreal.ca/"><img src="../assets/images/slides/logos/udem-white.png" alt="UdeM" style="height: 4em"></a> ] .center[ <a href="https://institut-courtois.umontreal.ca/"><img src="../assets/images/slides/logos/institut-courtois.png" alt="Institut Courtois" style="height: 2.5em"></a> <a href="https://ivado.ca/"><img src="../assets/images/slides/logos/ivado.png" alt="IVADO" style="height: 2.5em"></a> ] .smaller[.footer[ Slides: [alexhernandezgarcia.github.io/slides/{{ name }}](https://alexhernandezgarcia.github.io/slides/{{ name }}) ]] .qrcode[] --- count: false name: title class: title, middle ### Why scientific discoveries? .center[] --- ## Why scientific discoveries? .context[Climate change is a major challenge for humanity.] <br><br> .center[ <figure> <img src="../assets/images/slides/climatechange/ipcc_warming.png" alt="Observed (1900–2020) and projected (2021–2100) changes in global surface temperature (relative to 1850–1900)" style="width: 100%"> <figcaption>.smaller[Observed (1900–2020) and projected (2021–2100) changes in global surface temperature relative to 1850–1900 (adapted from: <a href="https://www.ipcc.ch/report/sixth-assessment-report-cycle/">IPCC Sixth Assessment Report</a>)]</figcaption> </figure> ] .conclusion["The evidence is clear: the time for action is now." .smaller[IPCC Report, 2022]] --- ## Why scientific discoveries? .context[Climate change is a major challenge for humanity.] .center[ <figure> <img src="../assets/images/slides/climatechange/who_climate_health.png" alt="Climate change presents a fundamental threat to human health." style="width: 100%"> <figcaption>.smaller[Climate-sensitive health risks (adapted from: <a href="https://www.who.int/news-room/fact-sheets/detail/climate-change-and-health">World Health Organization</a>)]</figcaption> </figure> ] .smaller[ * Environmental factors take the lives of around 13 million people _per year_. * Climate change affects people’s mental and physical health, access to clean air, safe water, food and health care. ] .full-width[ .conclusion["Climate change is the single biggest health threat facing humanity." .smaller[[WHO and WMO](https://climahealth.info/), 2024]] ] --- ## Why scientific discoveries? ### The potential of materials discovery .context["The time for action is now"] -- > "Limiting global warming will require major transitions in the energy sector. This will involve a substantial reduction in fossil fuel use, widespread electrification, .highlight1[improved energy efficiency, and use of alternative fuels (such as hydrogen)]." .cite[IPCC Sixth Assessment Report, 2022] > "Reducing industry emissions will entail coordinated action throughout value chains to promote all mitigation options, including demand management, .highlight1[energy and materials efficiency, circular material flows]." .cite[IPCC Sixth Assessment Report, 2022] -- <br> .conclusion[Mitigation of the climate crisis requires innovation in the materials sector.] ??? Antimicrobial resistance - https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance - https://www.who.int/news-room/feature-stories/detail/donors-making-a-difference--climate-change-and-its-impact-on-health - https://www.who.int/news/item/31-10-2022-who-and-wmo-launch-a-new-knowledge-platform-for-climate-and-health - https://www.who.int/news/item/08-02-2024-who-medically-important-antimicrobial-list-2024 - https://cdn.who.int/media/docs/default-source/gcp/who-mia-list-2024-lv.pdf?sfvrsn=3320dd3d_2 - https://www.who.int/publications/i/item/9789240047655 --- ## Why scientific discoveries? ### The potential of drug discovery .context[Drug discovery and vaccine development play a crucial role in modern healthcare systems.] .right-column-33[ .center[] ] --- count: false ## Why scientific discoveries? ### The potential of drug discovery .context[Drug discovery and vaccine development play a crucial role in modern healthcare systems.] .right-column-33[ .center[] ] .left-column-66[ .highlight1[Bacterial antimicrobial resistance] contributed to 4.95 million deaths in 2019. .cite[World Health Organisation (WHO), 2023] WHO's latest annual review identified 27 antibiotics in clinical development that address WHO bacterial priority pathogens, of which .highlight1[only 6 were classified as innovative]. "The recently approved antibacterial agents are .highlight1[insufficient to tackle the challenge] of increasing emergence and spread of antimicrobial resistance". .cite[World Health Organisation (WHO), 2021] ] --- count: false ## Why scientific discoveries? ### The potential of drug discovery .context[Drug discovery and vaccine development play a crucial role in modern healthcare systems.] .right-column-33[ .center[ <figure> <img src="../assets/images/slides/drugs/who_notimetowait.png" alt="No time to wait" style="width: 55%"> <figcaption><small>"No time to wait". Source: <a href="https://www.who.int/docs/default-source/documents/no-time-to-wait-securing-the-future-from-drug-resistant-infections-en.pdf">WHO</a>.</small></figcaption> </figure> ] ] .left-column-66[ .highlight1[Bacterial antimicrobial resistance] contributed to 4.95 million deaths in 2019. .cite[World Health Organisation (WHO), 2023] WHO's latest annual review identified 27 antibiotics in clinical development that address WHO bacterial priority pathogens, of which .highlight1[only 6 were classified as innovative]. "The recently approved antibacterial agents are .highlight1[insufficient to tackle the challenge] of increasing emergence and spread of antimicrobial resistance". .cite[World Health Organisation (WHO), 2021] ] .full-width[ .conclusion["No time to wait". Alongside other necessary actions, drug discovery plays a key role in tackling the antimicrobial resistance global threat.] ] --- ## Machine Learning for Science .center[] .conclusion[Machine learning research has the potential to facilitate scientific discoveries to tackle climate and health challenges.] --- count: false ## Machine Learning for Science and Science for Machine Learning .center[] .conclusion[Machine learning research has the potential to facilitate scientific discoveries to tackle climate and health challenges. Scientific challenges stimulate in turn machine learning research.] --- count: false name: mlforscience class: title, middle ### Machine learning for scientific discoveries .center[] --- ## Traditional discovery cycle .context35[The climate crisis demands accelerating scientific discoveries.] -- .right-column-66[<br>.center[]] .left-column-33[ <br> The .highlight1[traditional pipeline] for scientific discovery: * relies on .highlight1[highly specialised human expertise], * it is .highlight1[time-consuming] and * .highlight1[financially and computationally expensive]. ] --- count: false ## Machine learning in the loop .context35[The traditional scientific discovery loop is too slow for certain applications.] .right-column-66[<br>.center[]] .left-column-33[ <br> A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and ] --- count: false ## Machine learning in the loop .context35[The traditional scientific discovery loop is too slow for certain applications.] .right-column-66[<br>.center[]] .left-column-33[ <br> A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and * used to quickly and cheaply evaluate queries ] --- count: false ## Machine learning in the loop .context35[The traditional scientific discovery loop is too slow for certain applications.] .right-column-66[<br>.center[]] .left-column-33[ <br> A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and * used to quickly and cheaply evaluate queries .conclusion[There are infinitely many conceivable materials, $10^{180}$ potentially stable and $10^{60}$ drug molecules. Are predictive models enough?] ] --- count: false ## _Generative_ machine learning in the loop .right-column-66[<br>.center[]] .left-column-33[ <br> .highlight1[Generative machine learning] can: * .highlight1[learn structure] from the available data, * .highlight1[generalise] to unexplored regions of the search space and * .highlight1[build better queries] ] --- count: false ## _Generative_ machine learning in the loop .right-column-66[<br>.center[]] .left-column-33[ <br> .highlight1[Generative machine learning] can: * .highlight1[learn structure] from the available data, * .highlight1[generalise] to unexplored regions of the search space and * .highlight1[build better queries] .conclusion[Active learning with generative machine learning can in theory more efficiently explore the candidate space.] ] --- count: false name: title class: title, middle ### The challenges of scientific discoveries .center[] .center[] --- ## An intuitive trivial problem .highlight1[Problem]: find one arrangement of Tetris pieces on the board that minimise the empty space. .left-column-33[ .center[] ] .right-column-66[ .center[] ] -- .full-width[.center[ <figure> <img src="../assets/images/slides/tetris/mode1.png" alt="Score 12" style="width: 3%"> <figcaption>Score: 12</figcaption> </figure> ]] --- count: false ## An intuitive ~~trivial~~ easy problem .highlight1[Problem]: find .highlight2[all] the arrangements of Tetris pieces on the board that minimise the empty space. .left-column-33[ .center[] ] .right-column-66[ .center[] ] -- .full-width[.center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode1.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode2.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode3.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode4.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode5.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> </div> ]] --- count: false ## An intuitive ~~easy~~ hard problem .highlight1[Problem]: find .highlight2[all] the arrangements of Tetris pieces on the board that minimise the empty space. .left-column-33[ .center[] ] .right-column-66[ .center[] ] -- .full-width[.center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode1.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode2.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode3.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode4.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode5.png" alt="Random board" style="width: 40%"> </figure> </div> </div> ]] --- count: false ## An incredibly ~~intuitive easy~~ hard problem .highlight1[Problem]: find .highlight2[all] the arrangements of Tetris pieces on the board that .highlight2[optimise an unknown function]. .left-column-33[ .center[] ] .right-column-66[ .center[] ] -- .full-width[.center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_434.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_800.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_815.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_849.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_905.png" alt="Random board" style="width: 40%"> </figure> </div> </div> ]] --- count: false ## An incredibly ~~intuitive easy~~ hard problem .highlight1[Problem]: find .highlight2[all] the arrangements of Tetris pieces on the board that .highlight2[optimise an unknown function]. .left-column-33[ .center[] ] .right-column-66[ .center[] ] .full-width[.conclusion[Materials and drug discovery involve finding candidates with rare properties from combinatorially or infinitely many options.]] --- ## Why Tetris for scientific discovery? .context35[The "Tetris problem" involves .highlight1[sampling from an unknown distribution] in a .highlight1[discrete, high-dimensional, combinatorially large space].] --- count: false ## Why Tetris for scientific discovery? ### Biological sequence design <br> Proteins, antimicrobial peptides (AMP) and DNA can be represented as sequences of amino acids or nucleobases. There are $22^{100} \approx 10^{134}$ protein sequences with 100 amino acids. .context35[The "Tetris problem" involves sampling from an unknown distribution in a discrete, high-dimensional, combinatorially large space] .center[] -- .left-column-66[ .dnag[`G`].dnaa[`A`].dnag[`G`].dnag[`G`].dnag[`G`].dnac[`C`].dnag[`G`].dnaa[`A`].dnac[`C`].dnag[`G`].dnag[`G`].dnat[`T`].dnaa[`A`].dnac[`C`].dnag[`G`].dnag[`G`].dnaa[`A`].dnag[`G`].dnac[`C`].dnat[`T`].dnac[`C`].dnat[`T`].dnag[`G`].dnac[`C`].dnat[`T`].dnac[`C`].dnac[`C`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`]<br> .dnat[`T`].dnac[`C`].dnaa[`A`].dnac[`C`].dnac[`C`].dnat[`T`].dnac[`C`].dnac[`C`].dnac[`C`].dnag[`G`].dnaa[`A`].dnag[`G`].dnac[`C`].dnaa[`A`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnag[`G`].dnat[`T`].dnat[`T`].dnag[`G`].dnat[`T`].dnaa[`A`].dnag[`G`].dnag[`G`].dnac[`C`].dnaa[`A`].dnag[`G`].dnac[`C`].dnag[`G`].dnat[`T`].dnac[`C`].dnac[`C`].dnat[`T`].dnaa[`A`].dnac[`C`].dnac[`C`].dnag[`G`].dnat[`T`].dnat[`T`].dnac[`C`].dnag[`G`]<br> .dnac[`C`].dnat[`T`].dnaa[`A`].dnac[`C`].dnag[`G`].dnac[`C`].dnag[`G`].dnat[`T`].dnac[`C`].dnat[`T`].dnac[`C`].dnat[`T`].dnat[`T`].dnat[`T`].dnac[`C`].dnag[`G`].dnag[`G`].dnag[`G`].dnag[`G`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`]<br> .dnat[`T`].dnat[`T`].dnag[`G`].dnac[`C`].dnaa[`A`].dnag[`G`].dnaa[`A`].dnag[`G`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`].dnaa[`A`].dnac[`C`].dnag[`G`].dnac[`C`].dnag[`G`].dnac[`C`].dnaa[`A`].dnat[`T`].dnag[`G`].dnac[`C`].dnag[`G`].dnaa[`A`].dnac[`C`].dnat[`T`].dnag[`G`].dnag[`G`].dnag[`G`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`].dnag[`G`].dnat[`T`].dnaa[`A`].dnag[`G`].dnat[`T`].dnac[`C`].dnag[`G`].dnaa[`A`].dnaa[`A`].dnac[`C`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnat[`T`].dnat[`T`].dnag[`G`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnaa[`A`].dnaa[`A`].dnac[`C`].dnaa[`A`]<br> .dnag[`G`].dnac[`C`].dnat[`T`].dnac[`C`].dnag[`G`].dnac[`C`].dnat[`T`].dnat[`T`].dnaa[`A`].dnag[`G`].dnag[`G`].dnag[`G`].dnac[`C`].dnac[`C`].dnat[`T`].dnac[`C`].dnag[`G`].dnaa[`A`].dnac[`C`].dnat[`T`].dnac[`C`].dnac[`C`].dnat[`T`].dnac[`C`].dnat[`T`].dnag[`G`].dnaa[`A`].dnaa[`A`].dnat[`T`].dnag[`G`].dnag[`G`].dnaa[`A`].dnag[`G`].dnat[`T`].dnag[`G`].dnat[`T`].dnat[`T`].dnac[`C`].dnaa[`A`].dnat[`T`].dnac[`C`].dnag[`G`].dnaa[`A`].dnaa[`A`].dnat[`T`].dnag[`G`].dnag[`G`].dnaa[`A`].dnag[`G`].dnat[`T`].dnag[`G`]<br> ] --- ## Why Tetris for scientific discovery? ### Molecular generation .context35[The "Tetris problem" involves sampling from an unknown distribution in a discrete, high-dimensional, combinatorially large space] <br> Small molecules can also be represented as sequences or by a combination of of higher-level fragments. There may be about $10^{60}$ drug-like molecules. -- .columns-3-left[ .center[  `CC(=O)NCCC1=CNc2c1cc(OC)cc2 CC(=O)NCCc1c[nH]c2ccc(OC)cc12` ]] .columns-3-center[ .center[  `OCCc1c(C)[n+](cs1)Cc2cnc(C)nc2N` ]] .columns-3-right[ .center[  `CN1CCC[C@H]1c2cccnc2` ]] --- ## Why Tetris for scientific discovery? ### Crystal structure generation .context35[The "Tetris problem" involves sampling from an unknown distribution in a discrete, high-dimensional, combinatorially large space] <br> Crystal structures can be described by their chemical composition, the symmetry group and the lattice parameters (and more generally by atomic positions). -- .center[] .references[ * Mila AI4Science et al. [Crystal-GFN: sampling crystals with desirable properties and constraints](https://arxiv.org/abs/2310.04925). AI4Mat, NeurIPS 2023 (spotlight). ] --- ## Machine learning for scientific discovery ### Challenges and limitations of existing methods -- .highlight1[Challenge]: very large and high-dimensional search spaces. -- → Need for .highlight2[efficient search and generalisation] of underlying structure. -- .highlight1[Challenge]: underspecification of objective functions or metrics. -- → Need for .highlight2[diverse] candidates. -- .highlight1[Limitation]: Reinforcement learning excels at optimisation in complex spaces but tends to lack diversity. -- .highlight1[Limitation]: Markov chain Monte Carlo (MCMC) can _sample_ from a distribution (diversity) but struggles at mode mixing in high dimensions. -- → Need to .highlight2[combine all of the above]: sampling from complex, high-dimensional distributions. -- .conclusion[Generative flow networks (GFlowNets) and active learning address these challenges.] --- count: false name: relatedwork class: title, middle ## Related approaches ### Could we use off-the-shelf algorithms? .center[] --- ## Related work ### Bayesian optimisation .context[What are the most relevant _searching algorithms_?] Definition: Bayesian optimization is a sequential design strategy for _global optimization_ of black-box functions, that does not assume any functional forms. .cite[[Wikipedia, Feb. 2025](https://en.wikipedia.org/wiki/Bayesian_optimization)] .center[ $$x^{\star} = \text{arg max} f(x)$$ ] .center[ <figure> <img src="../assets/images/slides/activelearning/objective_function.png" alt="Bayesian optimisation" style="width: 40%"> <figcaption><small>Source: <a href="https://bayesoptbook.com/">Roman Garnett. Bayesian Optimization. Cambridge University Press, 2023</a>.</small></figcaption> </figure> ] .conclusion[Bayesian optimisation is not concerned with discovering multiple high-scoring data points, but it offers a suitable framework as starting point.] --- ## Related work ### Active search .context[What are the most relevant _searching algorithms_?] Definition: Given a search space with data points belonging to two classes, active search is the problem of locating the members of one particular class as quickly as possible. .cite[(Garnett et al., 2012)] .left-column[ Given a set of observations $\mathcal{D} \triangleq {(x_i, y_i)}$, active search aims to optimise the utility function defined as the number of targets found: $u(\mathcal{D}) \triangleq \sum y_i$. ] .right-column[ .center[ <figure> <img src="../assets/images/slides/activelearning/active_search.png" alt="Active search" style="width: 60%"> </figure> ] ] .references[ * Garnett et al. [Bayesian optimal active search and surveying](Bayesian optimal active search and surveying). ICML 2012. ] --- count: false ## Related work ### Active search .context[What are the most relevant _searching algorithms_?] Definition: Given a search space with data points belonging to two classes, active search is the problem of locating the members of one particular class as quickly as possible. .cite[(Garnett et al., 2012)] .left-column[ Given a set of observations $\mathcal{D} \triangleq {(x_i, y_i)}$, active search aims to optimise the utility function defined as the number of targets found: $u(\mathcal{D}) \triangleq \sum y_i$. ] .right-column[ .center[ <figure> <img src="../assets/images/slides/activelearning/active_search.png" alt="Active search" style="width: 60%"> </figure> ] ] .conclusion[Bayesian active search is interesting for materials and drug discovery but it reduces the value of candidates to binary classes.] --- ## Related work ### Quality Diversity .context[What are the most relevant _searching algorithms_?] Definition: A class of evolutionary algorithms which puts emphasis on diversity while searching for optimal or near optimal solutions on a latent space. .center[ <figure> <img src="../assets/images/slides/activelearning/mapelites.png" alt="MAP Elites" style="width: 60%"> <figcaption><small>Source: <a href="https://arxiv.org/abs/1504.04909">Mouret and Clune. Illuminating search spaces by mapping elites. 2015</a>.</small></figcaption> </figure> ] .conclusion[Quality Diversity (QD) algorithms share the objective of finding diverse, high-scoring candidates, despite emerging from a different research community. Definitely something to try soon too.] --- ## Related work ### Active learning .context[What are the most relevant _searching algorithms_?] Definition: A class of machine learning methods whose goal is to learn an efficient data sampling scheme to accelerate training. .center[ <figure> <img src="../assets/images/slides/activelearning/activelearning_settles.png" alt="Active learning" style="width: 60%"> <figcaption><small>Source: <a href="https://burrsettles.com/pub/settles.activelearning.pdf">Burr Settles. Active learning literature survey. Independent Technical Report, 2009</a>.</small></figcaption> </figure> ] .conclusion[Active learning is a large family of algorithms or problems that includes our own. However, most of the literature has focused on _pool-based active learning_.] ??? Mention: - Multi-armed bandits - Experimental design - The review in Jain et al. --- count: false name: mfal class: title, middle ## Multi-fidelity active learning Nikita Saxena, Moksh Jain, Cheng-Hao Liu, Yoshua Bengio .smaller[[Multi-fidelity active learning with GFlowNets](https://arxiv.org/abs/2306.11715). Transactions on Machine Learning Research (TMLR). 2024.] .center[] --- ## Why multi-fidelity? .context35[We had described the scientific discovery loop as a cycle with one single oracle.] <br><br> .right-column[ .center[] ] -- .left-column[ Example: "incredibly hard" Tetris problem: find arrangements of Tetris pieces that optimise an .highlight2[unknown function $f$]. - $f$: Oracle, cost per evaluation 1000 CAD. .center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_434.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_800.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_815.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_849.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_905.png" alt="Random board" style="width: 200%"> </figure> </div> </div> ] ] --- count: false ## Why multi-fidelity? .context35[However, in practice, multiple oracles (models) of different fidelity and cost are available in scientific applications.] <br><br> .right-column[ .center[] ] .left-column[ Example: "incredibly hard" Tetris problem: find arrangements of Tetris pieces that optimise an .highlight2[unknown function $f$]. - $f$: Oracle, cost per evaluation 1000 CAD. .center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_434.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_800.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_815.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_849.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_905.png" alt="Random board" style="width: 200%"> </figure> </div> </div> ] ] --- count: false ## Why multi-fidelity? .context35[However, in practice, multiple oracles (models) of different fidelity and cost are available in scientific applications.] <br><br> .right-column[ .center[] ] .left-column[ Example: "incredibly hard" Tetris problem: find arrangements of Tetris pieces that optimise an .highlight2[unknown function $f$]. - $f$: Oracle, cost per evaluation 1000 CAD. - $f\_1$: Slightly inaccurate oracle, cost 100 CAD. - $f\_2$: Noisy but informative oracle, cost 1 CAD. .center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_434.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_800.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_815.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_849.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_905.png" alt="Random board" style="width: 200%"> </figure> </div> </div> ] ] --- count: false ## Why multi-fidelity? .context[In many scientific applications we have access to multiple approximations of the objective function.] .left-column[ For example, in .highlight1[material discovery]: * .highlight1[Synthesis] of a material and characterisation of a property in the lab * Molecular dynamic .highlight1[simulations] to estimate the property * .highlight1[Machine learning] models trained to predict the property ] .right-column[ .center[] ] -- .conclusion[However, current machine learning methods cannot efficiently leverage the availability of multiple oracles and multi-fidelity data. Especially with .highlight1[structured, large, high-dimensional search spaces].] --- ## Contribution - An .highlight1[active learning] algorithm to leverage the availability of .highlight1[multiple oracles at different fidelities and costs]. -- - The goal is two-fold: 1. Find high-scoring candidates 2. Candidates must be diverse -- - Experimental evaluation with .highlight1[biological sequences and molecules]: - DNA - Antimicrobial peptides - Small molecules - Classical multi-fidelity toy functions (Branin and Hartmann) -- .conclusion[Likely the first multi-fidelity active learning method for biological sequences and molecules.] --- ## Our active learning algorithm .center[] --- count: false ## Our active learning algorithm .center[] --- count: false ## Our active learning algorithm .center[] --- count: false ## Our active learning algorithm .center[] --- count: false ## Our active learning algorithm .center[] --- count: false ## Our active learning algorithm .center[] --- count: false ## Our active learning algorithm .center[] --- count: false ## Our active learning algorithm .center[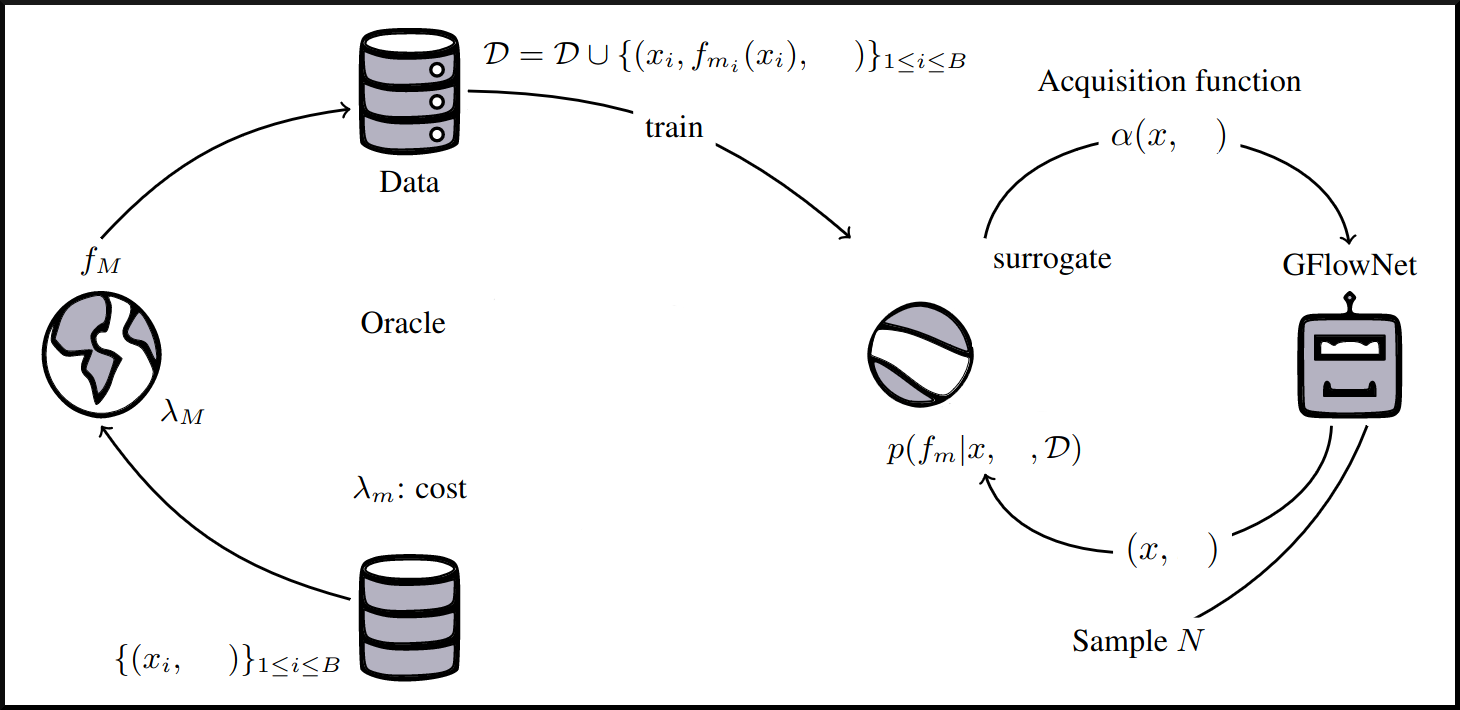] --- count: false ## Our active learning algorithm .center[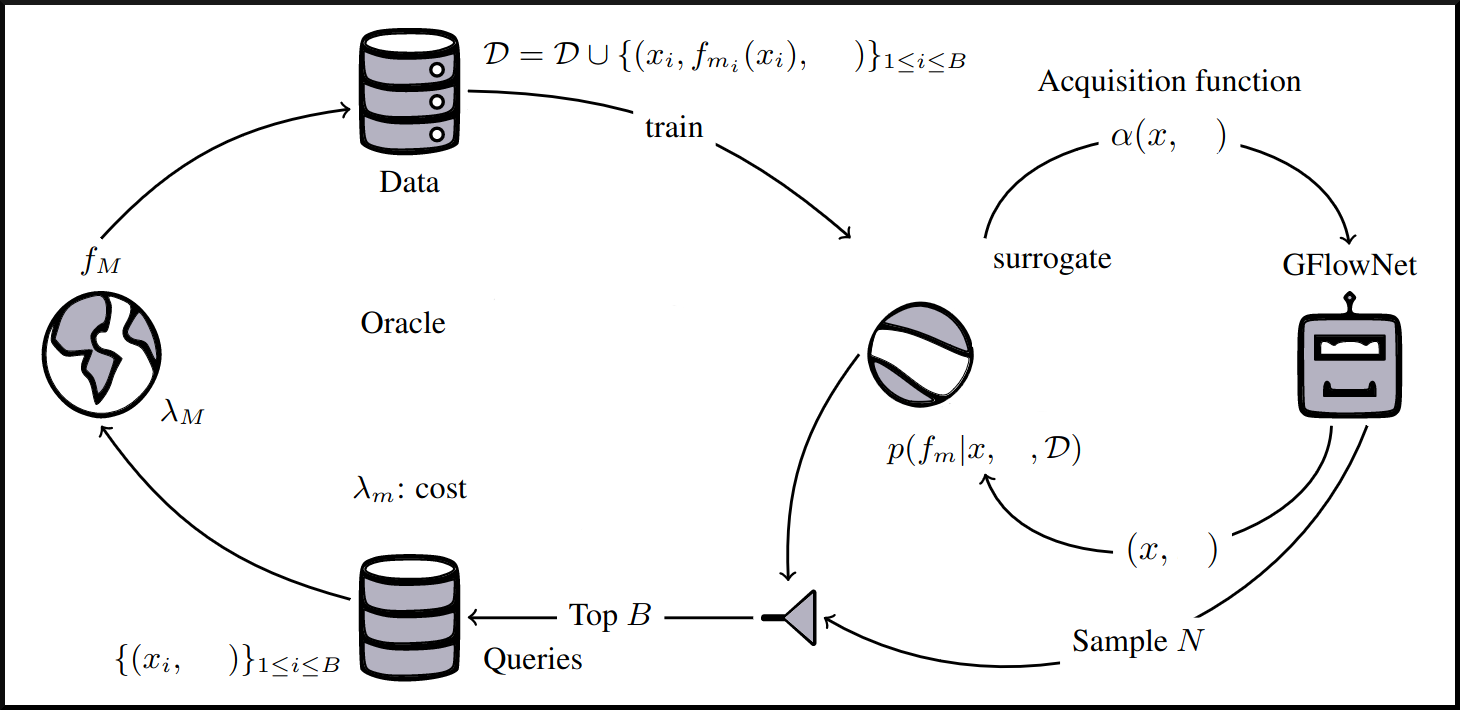] --- count: false ## Our multi-fidelity active learning algorithm .center[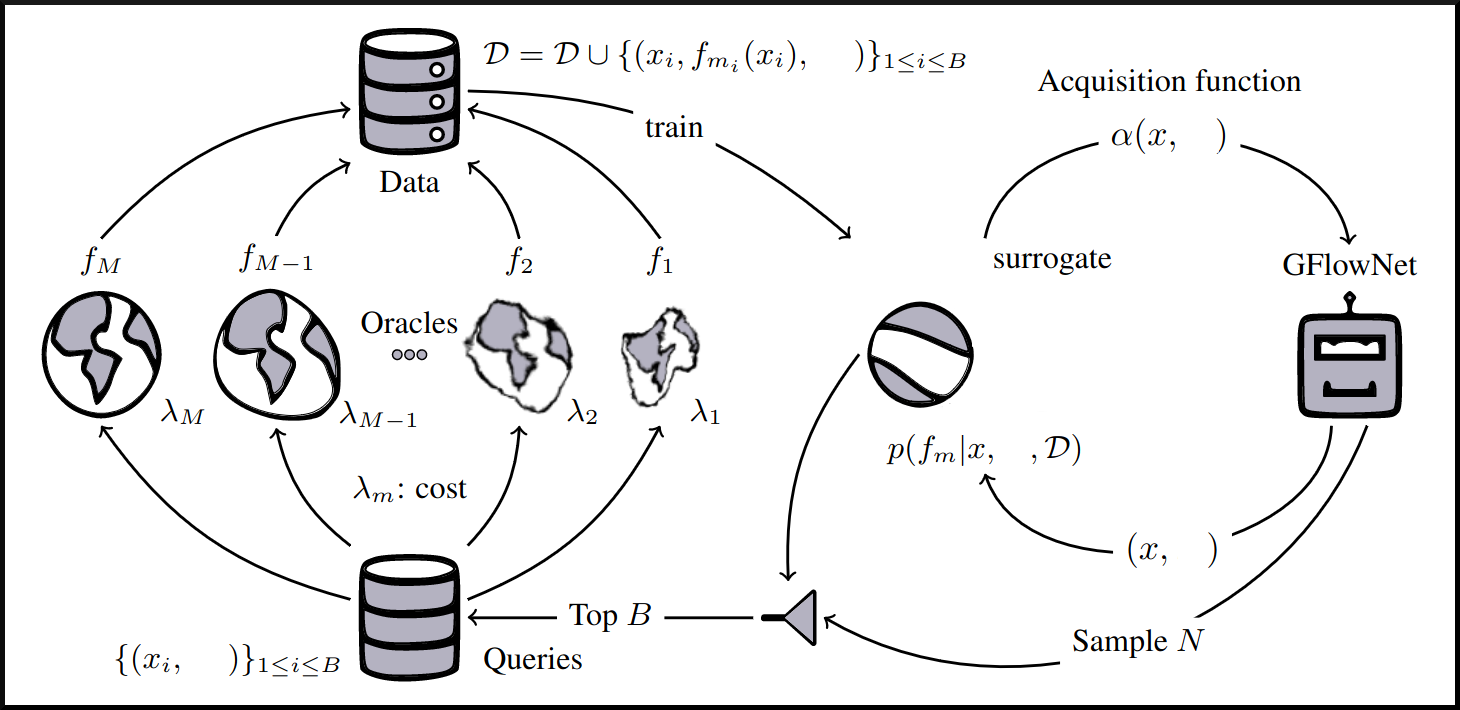] --- count: false ## Our multi-fidelity active learning algorithm .center[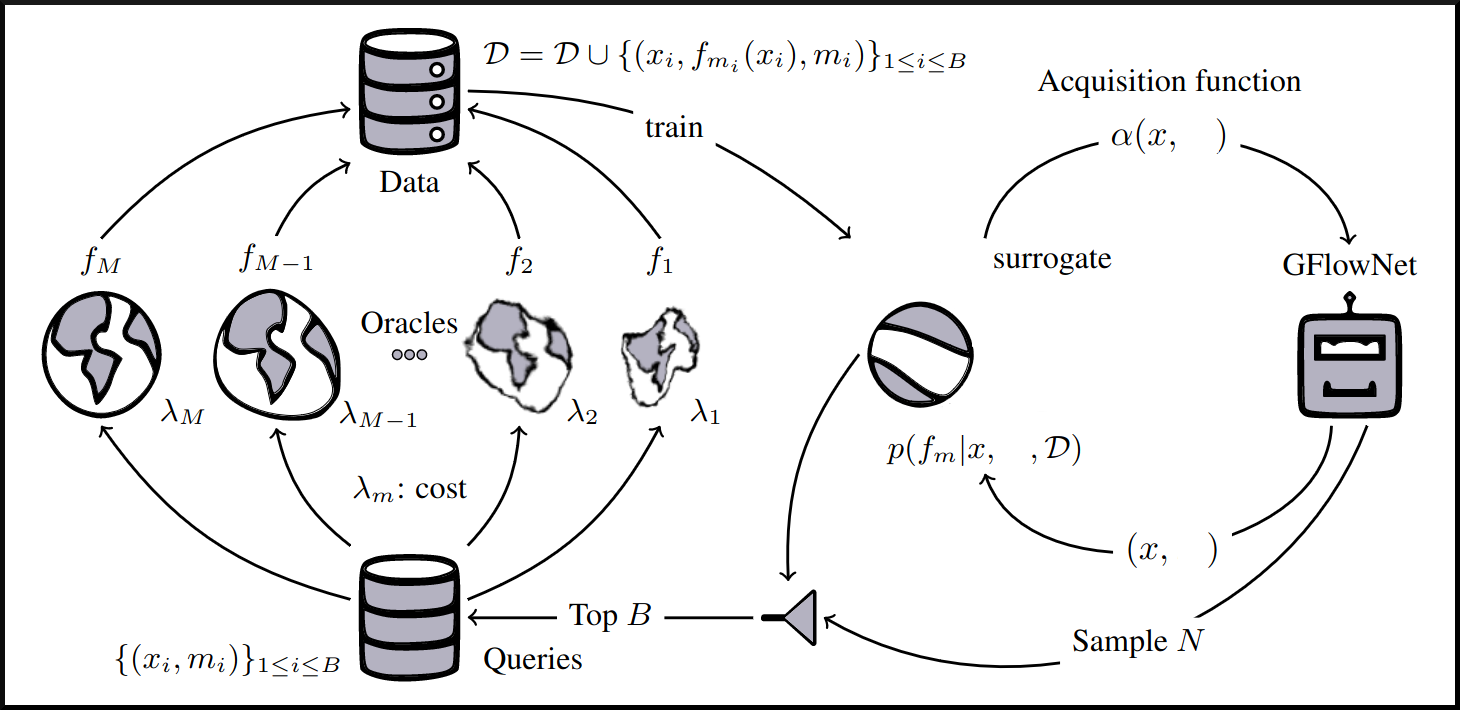] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- ## Experiments ### Baselines .context[This may be the .highlight1[first multi-fidelity active learning algorithm tested on biological sequence design and molecular design problems]. There did not exist baselines from the literature.] -- <br> * .highlight1[SF-GFN]: GFlowNet with highest fidelity oracle to establish a benchmark for performance without considering the cost-accuracy trade-offs. -- * .highlight1[Random]: Quasi-random approach where the candidates and fidelities are picked randomly and the top $(x, m)$ pairs scored by the acquisition function are queried. -- * .highlight1[Random fid. GFN]: GFlowNet with random fidelities, to investigate the benefit of deciding the fidelity with GFlowNets. -- * .highlight1[MF-PPO]: Replacement of MF-GFN with a reinforcement learning algorithm to _optimise_ the acquisition function. --- ## Small molecules - Realistic experiments with experimental oracles and costs that reflect computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. -- .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic .highlight1[ionisation potential (IP)]. Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- ## DNA aptamers - GFlowNet adds one nucleobase (`A`, `T`, `C`, `G`) at a time up to length 30. This yields a design space of size $|\mathcal{X}| = 4^{30}$. - The objective function is the free energy estimated by a bioinformatics tool. - The (simulated) lower fidelity oracle is a transformer trained with 1 million sequences. -- .center[] --- count: false ## Antimicrobial peptides (AMP) - Protein sequences (20 amino acids) with variable length (max. 50). - The oracles are 3 ML models trained with different subsets of data. -- .center[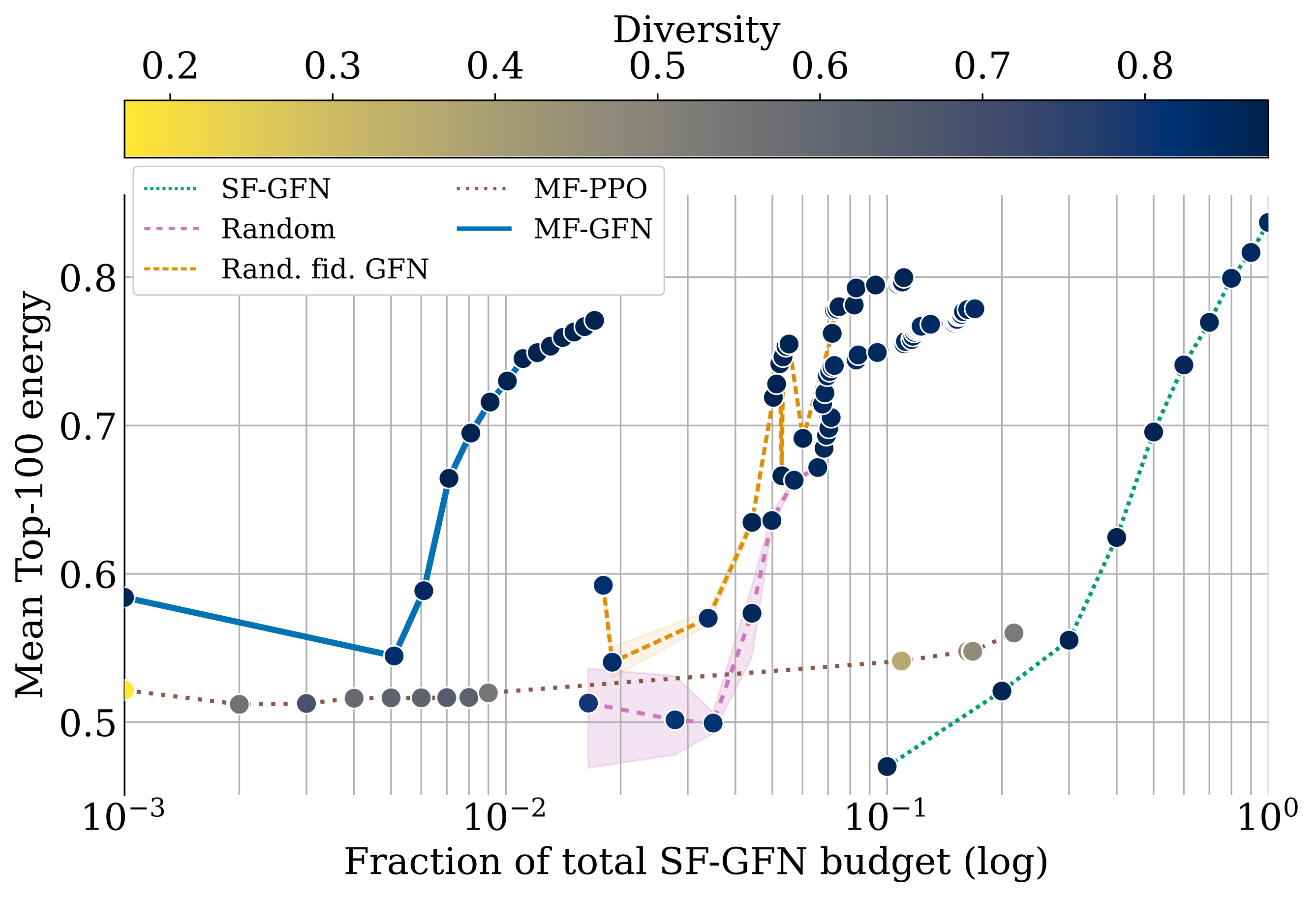] --- ## How does multi-fidelity help? .context[Visualisation on the synthetic 2D Branin function task.] .center[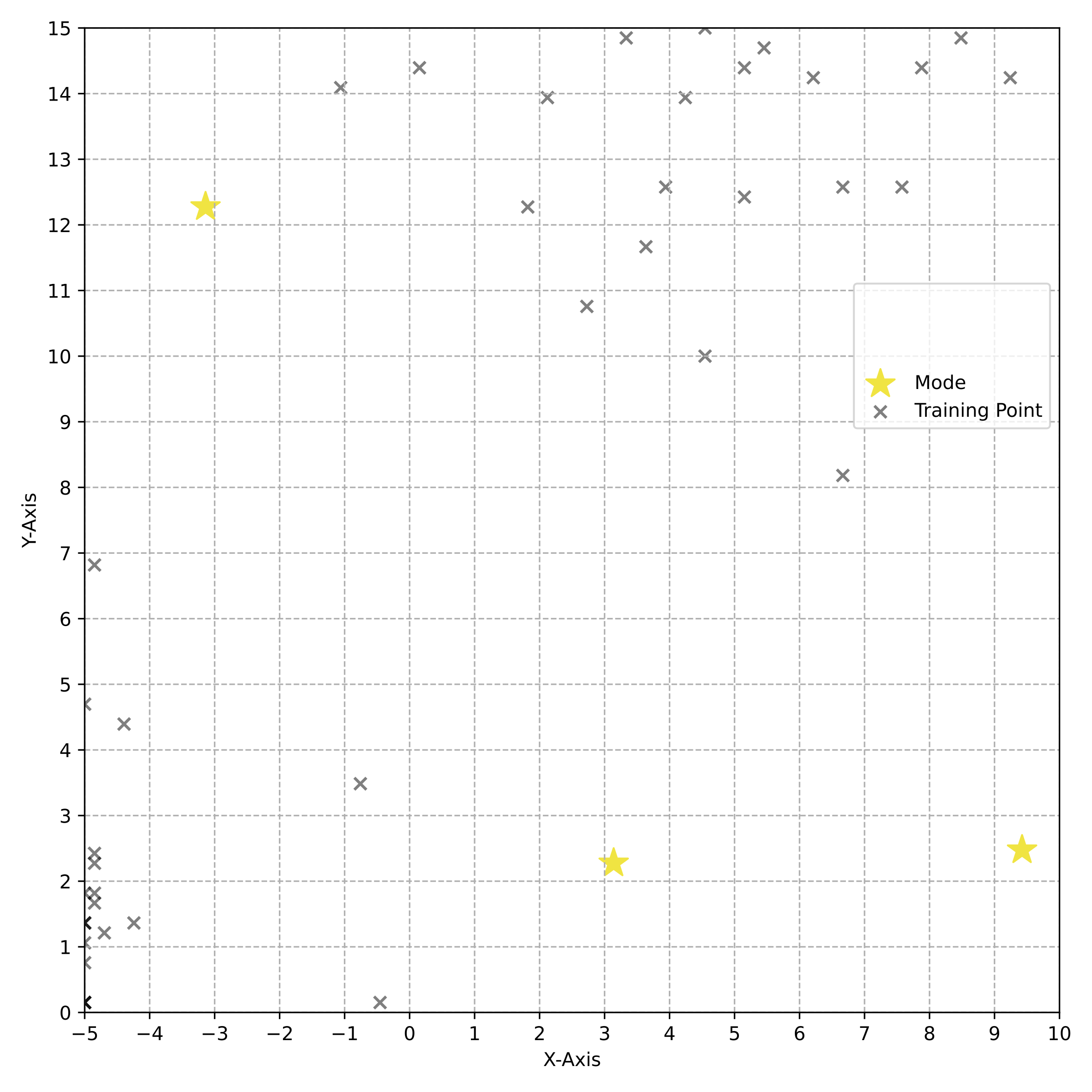] --- count: false ## How does multi-fidelity help? .context[Visualisation on the synthetic 2D Branin function task.] .center[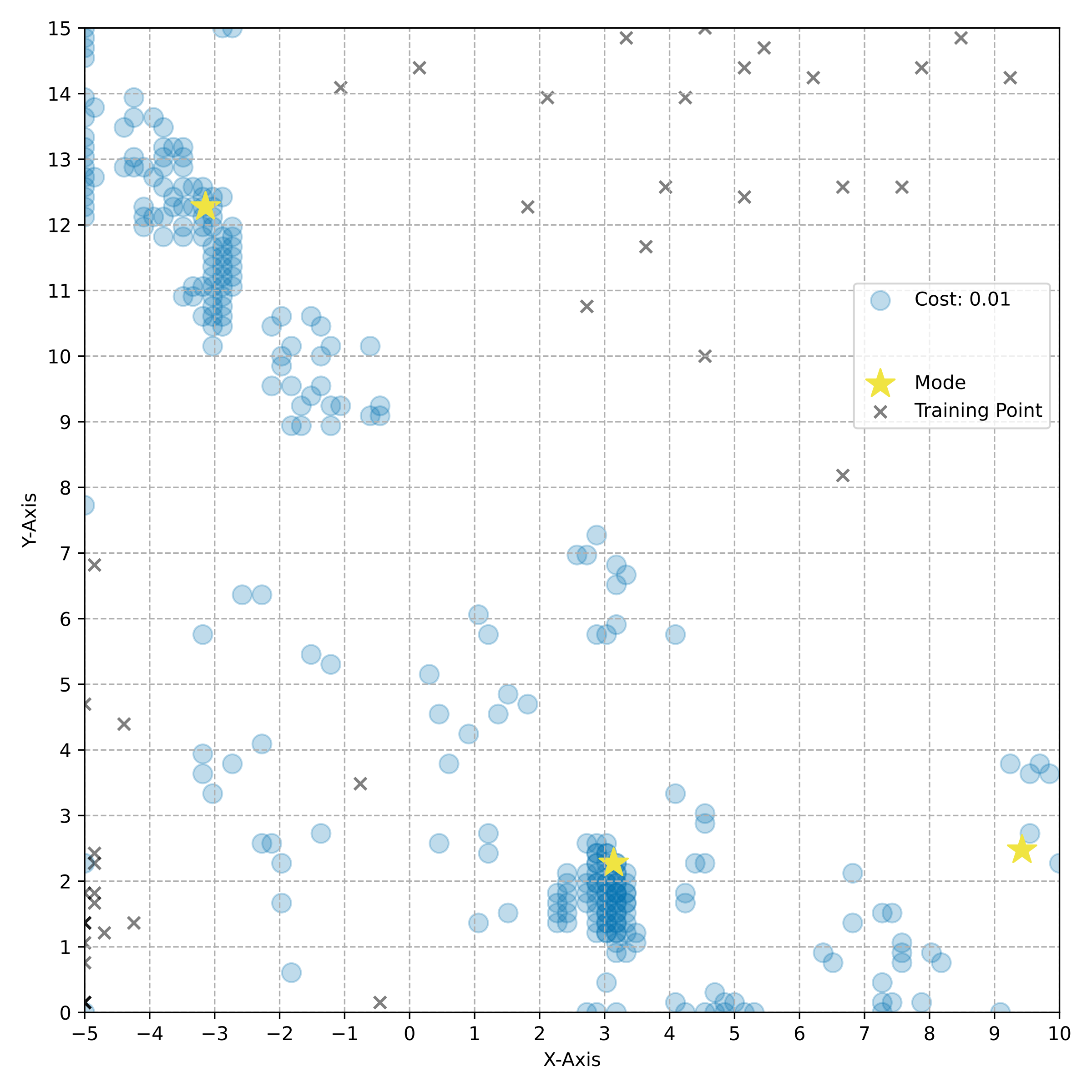] --- count: false ## How does multi-fidelity help? .context[Visualisation on the synthetic 2D Branin function task.] .center[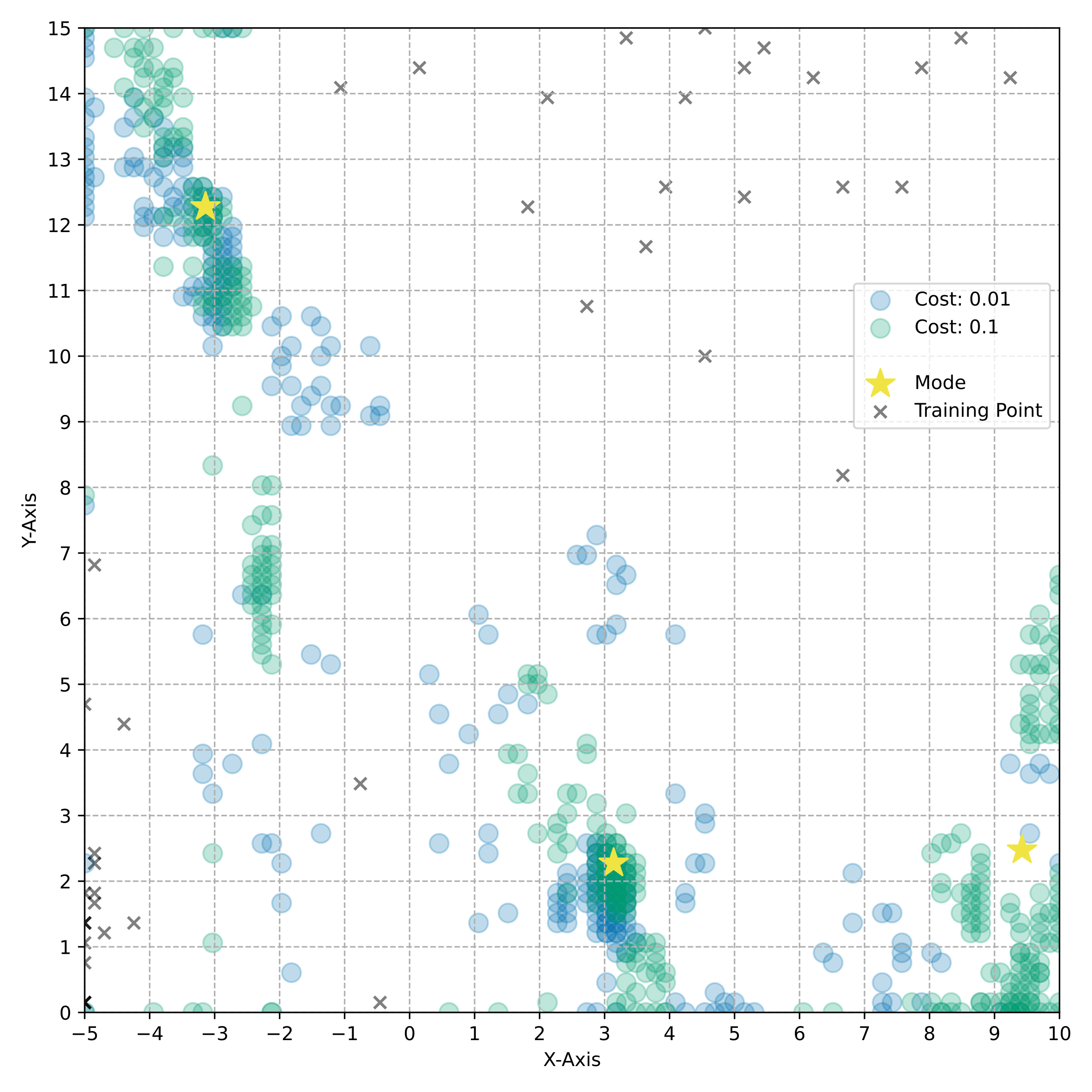] --- count: false ## How does multi-fidelity help? .context[Visualisation on the synthetic 2D Branin function task.] .center[] --- ## Details of the algorithm ### Multi-fidelity surrogate models * Small (synthetic) tasks: exact Gaussian Processes * Larger-scale, benchmark tasks: Deep Kernel Learning with stochastic variational Gaussian processes Multi-fidelity kernel learning: $$K_{MF}((x, m), (\tilde{x}, \tilde{m})) = K_X(g(x), g(\tilde{x})) + K_M(m, \tilde{m}) \times K_X^M(g(x), g(\tilde{x}))$$ * $K_X$ and $K_X^M$: Matérn kernels with different lengthscales each * Kernel of the fidelity confidences: $K_M(i, j) = (1 - \ell_i)(1 - \ell_j)(1 + \ell_i\ell_j)$ * $\ell_m = \frac{\lambda_m}{\lambda_M}$ .references[ * Wilson, Hu et al. [Deep Kernel Learning](https://arxiv.org/abs/1511.02222), AISTATS, 2016. * Mikkola et al. [Multi-fidelity Bayesian optimization with unreliable information sources](https://arxiv.org/abs/2210.13937) , AISTATS, 2023. ] --- ## Details of the algorithm ### Multi-fidelity acquisition function: Maximum Entropy Search (MES) MES it aims to maximise the mutual information between .hihglight1[the value] of the objective function $f$ when choosing point *x* and the maximum of the objective function, $f^{\star}$ (instead of considering the `arg max`). The multi-fidelity variant is designed to select the candidate $x$ and the fidelity $m$ that maximise the mutual information between $f_M^\star$ and the oracle at fidelity $m$, $f_m$ , weighted by the cost of the oracle $\lambda_m$. $$\alpha(x, m) = \frac{1}{\lambda_{m}} I(f_M^\star; f_m(x) | \mathcal{D})$$ .references[ * Moss et al. [GIBBON: General-purpose Information-Based Bayesian OptimisatioN](https://arxiv.org/abs/2102.03324), JMLR, 2021. ] --- ## Details of the algorithm ### Multi-fidelity GFlowNets (MF-GFN) Given a baseline GFlowNet with state space $\mathcal{S}$ and action space $\mathcal{A}$, we augment the state space with a new dimension for the fidelity $\mathcal{M'} = \{0, 1, 2, \ldots, M\}$ (including $m = 0$, which corresponds to unset fidelity): $\mathcal{S}_M = \mathcal{S} \times \mathcal{M'}$ The set of allowed transitions $\mathcal{A}_M$ is augmented such that a fidelity $m > 0$ of a trajectory must be selected once, and only once, from any intermediate state. This is meant to provide flexibility and improve generalisation. Finished trajectories are the concatenation of an object $x$ and the fidelity $m$: $(x, m) \in \mathcal{X}_M = \mathcal{X} \times \mathcal{M}$. GFlowNet is trained with the acquisition function $\alpha(x, m)$ as reward function. --- ## Applications ### Ongoing, planned and potential * Discovering materials with high ionic conductivity for solid-state electrolyte batteries. * Discovering novel antibiotics through a lab-in-the-loop approach. * Designing electrocatalysts for sustainability purposes. * Designing DNA aptamers and proteins that can bind to specific targets. * `<your application here>` --- ## Multi-fidelity active learning with GFlowNets ### Summary and conclusions .references[ * Hernandez-Garcia, Saxena et al. [Multi-fidelity active learning with GFlowNets](https://arxiv.org/abs/2306.11715). Transactions on Machine Learning Research (TMLR). 2024. ] * Current ML for science methods do not utilise all the information and resources at our disposal. -- * AI-driven scientific discovery demands learning methods that can .highlight1[efficiently discover diverse candidates in combinatorially large, high-dimensional search spaces]. -- * .highlight1[Multi-fidelity active learning with GFlowNets] enables .highlight1[cost-effective exploration] of large, high-dimensional and structured spaces, and discovers multiple, diverse modes of black-box score functions. -- * This is to our knowledge the first algorithm capable of effectively leveraging multi-fidelity oracles to discover diverse biological sequences and molecules. -- * .highlight2[Open source code]: * [github.com/nikita-0209/mf-al-gfn](https://github.com/nikita-0209/mf-al-gfn) * [github.com/alexhernandezgarcia/gflownet](https://github.com/alexhernandezgarcia/gflownet) --- ## Acknowledgements .columns-4[ .center[] .center[Nikita Saxena] ] .columns-4[ .center[] .center[Moksh Jain] ] .columns-4[ .center[] .center[Chenghao Liu] ] .columns-4[ .center[] .center[Yoshua Bengio] ] --- name: tea-talk-feb25 class: title, middle  Alex Hernández-García (he/il/él) .center[ <a href="https://mila.quebec/"><img src="../assets/images/slides/logos/mila-beige.png" alt="Mila" style="height: 3em"></a> <a href="https://www.umontreal.ca/"><img src="../assets/images/slides/logos/udem-white.png" alt="UdeM" style="height: 3em"></a> <a href="https://institut-courtois.umontreal.ca/"><img src="../assets/images/slides/logos/institut-courtois.png" alt="Institut Courtois" style="height: 3em"></a> <a href="https://ivado.ca/"><img src="../assets/images/slides/logos/ivado.png" alt="IVADO" style="height: 3em"></a> ] .highlight2[We are looking for students and collaborators to work on multi-fidelity active learning!] .footer[[alexhernandezgarcia.github.io](https://alexhernandezgarcia.github.io/) | [alex.hernandez-garcia@mila.quebec](mailto:alex.hernandez-garcia@mila.quebec)] | [alexhergar.bsky.social](https://bsky.app/profile/alexhergar.bsky.social) [](https://bsky.app/profile/alexhergar.bsky.social)<br> .smaller[.footer[ Slides: [alexhernandezgarcia.github.io/slides/{{ name }}](https://alexhernandezgarcia.github.io/slides/{{ name }}) ]]