name: title class: title, middle ## Deep active learning for DNA aptamer design Alex Hernández-García (he/il/él) .turquoise[Samsung-Mila-NYU Workshop · Online · Nov. 3rd 2021] .center[ <a href="https://mila.quebec/"><img src="../assets/images/slides/logos/mila-beige.png" alt="Mila" style="height: 4em"></a> <a href="http://wimlds.org/"><img src="../assets/images/slides/logos/udem-white.png" alt="UdeM" style="height: 4em"></a> ] .footer[[alexhernandezgarcia.github.io](https://alexhernandezgarcia.github.io/) | [alex.hernandez-garcia@mila.quebec](mailto:alex.hernandez-garcia@mila.quebec) | [@alexhdezgcia](https://twitter.com/alexhdezgcia)] [](https://twitter.com/alexhdezgcia) --- ## Collaboration .center[  ] * .bigger[[Michael Kilgour](https://sites.google.com/view/michael-kilgour/home)] (**lead**), Postdoc at NYU, formerly McGill * Danny Salem, University of Ottawa * Tao Liu, McGill * [Miroslava Cuperlovic-Culf](https://med.uottawa.ca/bmi/en/people/cuperlovic-culf-miroslava), University of Ottawa * [Yoshua Bengio](https://yoshuabengio.org/), Professor at UdeM and Mila * [Lena Simine](https://www.mcgill.ca/chemistry/faculty/lena-simine), Professor at McGill, Department of Chemistry .center[ <a href="https://mila.quebec/"><img src="../assets/images/slides/logos/mila-beige.png" alt="Mila" style="height: 3em"></a> <a href="http://wimlds.org/"><img src="../assets/images/slides/logos/udem-white.png" alt="UdeM" style="height: 3em"></a> <a href="http://wimlds.org/"><img src="../assets/images/slides/logos/mcgill.png" alt="McGill" style="height: 3em"></a> ] --- ## Motivation ### DNA aptamers .context[What are DNA aptamers and why do we care?] -- * DNA aptamers are short, single-stranded nucleotide (ssDNA) sequences. Example: `TATGCATGTGGGCGACGCAGTGCCCGTGGGATTTACTTGCAC` -- * DNA aptamers have multiple applications as .highlight1[aptasensors] .cite[([Kilgour et al., 2021](https://chemrxiv.org/engage/chemrxiv/article-details/60cbcb1d461f5627524764ab))]: * Antibiotics .cite[([Mehlhorn et al., 2018](https://www.mdpi.com/2079-6374/8/2/54))] * Neurotransmitters .cite[([Sinha et al., 2020](https://sci-hub.st/https://link.springer.com/article/10.1007%2Fs12038-020-0017-x))] * Steroids .cite[([Ebrahimi et al., 2021](https://sci-hub.st/https://onlinelibrary.wiley.com/doi/epdf/10.1002/anie.202103440))] * Metals .cite[([Zhou et al., 2017](https://pubs.acs.org/doi/10.1021/acs.chemrev.7b00063))] * Proteins .cite[([Kirby et al., 2004](https://sci-hub.st/https://pubs.acs.org/doi/pdf/10.1021/ac049858n))] * Adenosine triphosphate .cite[([Huizenga et al., 1995](https://sci-hub.st/10.1021/bi00002a033))] -- * Aptasensors are .highlight1[stable and selective] in crowded biochemical environments. -- * Designing aptamers that selectively and reliably bind a target analyte is highly non-trivial. -- .conclusion[Improving the pipeline for DNA aptamers design can potentially impact a wide range of applications.] --- ## Motivation ### DNA aptamer design .context[The traditional pipeline] .right-column-66[.center[]] .left-column-33[ The .highlight1[traditional pipeline] for DNA aptamer design (as other domains of scientific discovery): * relies on .highlight1[highly specialised human expertise], * it is .highlight1[time-consuming] and * .highlight1[financially and computationally expensive]. ] --- count: false ## Motivation ### DNA aptamer design .context[Machine learning in the loop] .right-column-66[.center[]] .left-column-33[ A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and * used to quickly and cheaply evaluate queries ] --- count: false ## Motivation ### DNA aptamer design .context[Machine learning in the loop] .right-column-66[.center[]] .left-column-33[ A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and * used to quickly and cheaply evaluate queries .conclusion[A machine learning model replacing real-world experiments can _only_ provide _linear_ gains.] ] --- count: false ## Motivation ### DNA aptamer design .context[Can we do better than _linear_? An agent in the loop] .right-column-66[.center[]] .left-column-33[ A .highlight1[machine learning **agent**] in the loop can: * .highlight1[learn structure] from the available data, * .highlight1[generalise] to unexplored regions of the search space and * .highlight1[build better queries] ] --- count: false ## Motivation ### DNA aptamer design .context[Can we do better than _linear_? An agent in the loop] .right-column-66[.center[]] .left-column-33[ A .highlight1[machine learning **agent**] in the loop can: * .highlight1[learn structure] from the available data, * .highlight1[generalise] to unexplored regions of the search space and * .highlight1[build better queries] .conclusion[A successful active learning pipeline with an ML agent in the loop can provide _exponential_ gains.] ] --- ## Methodology ### Overview .context[An **active learning** pipeline for DNA aptamer design] -- .right-column-66[.center[]] --- count: false ## Methodology ### Overview .context[An **active learning** pipeline for DNA aptamer design] .right-column-66[.center[]] --- count: false ## Methodology ### Overview .context[An **active learning** pipeline for DNA aptamer design] .right-column-66[.center[]] .left-column-33[ **Oracle**: [NUPACK](http://www.nupack.org/), a Python package for the analysis of the minimum free-energy structure of DNA and RNA sequences. .center[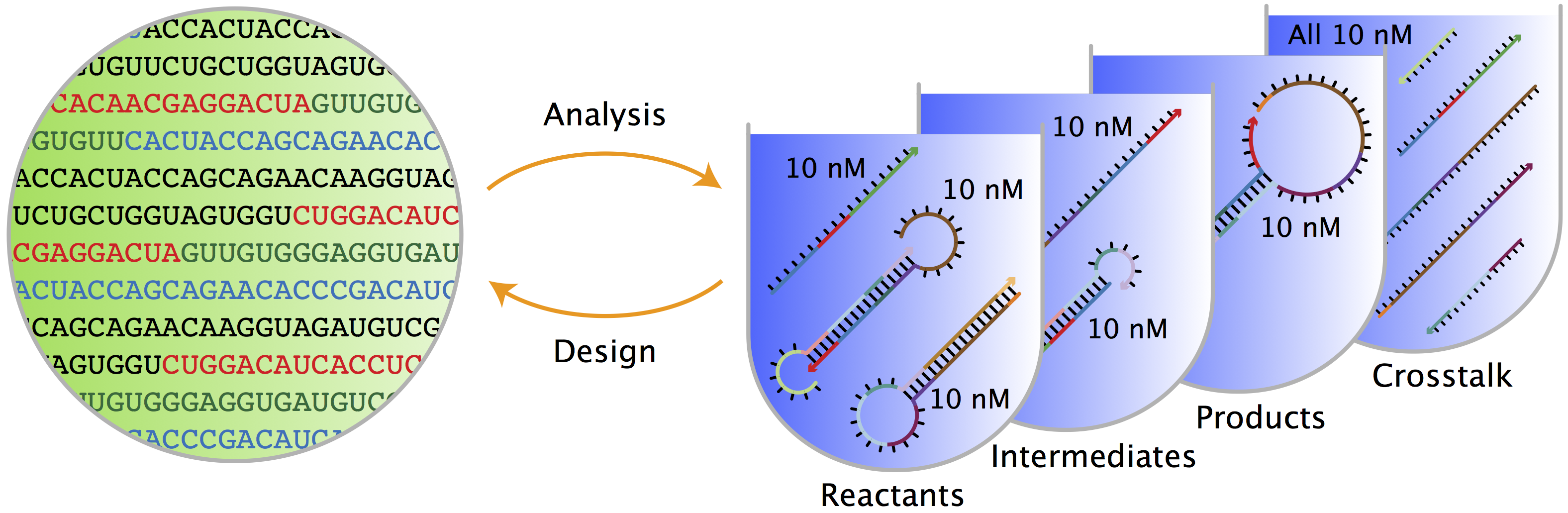] Used as a proxy oracle for developing and testing the active learning pipeline. ] --- count: false ## Methodology ### Overview .context[An **active learning** pipeline for DNA aptamer design] .right-column-66[.center[]] .left-column-33[ **Model**: a machine learning algorithm: multilayer perceptrons and transformers. ] --- count: false ## Methodology ### Overview .context[An **active learning** pipeline for DNA aptamer design] .right-column-66[.center[]] .left-column-33[ **GFlowNet**: a generative model that learns to produce diverse candidates with a probability proportional to their reward .cite[[(Bengio et al., 2021)](https://arxiv.org/abs/2106.04399)]. .center[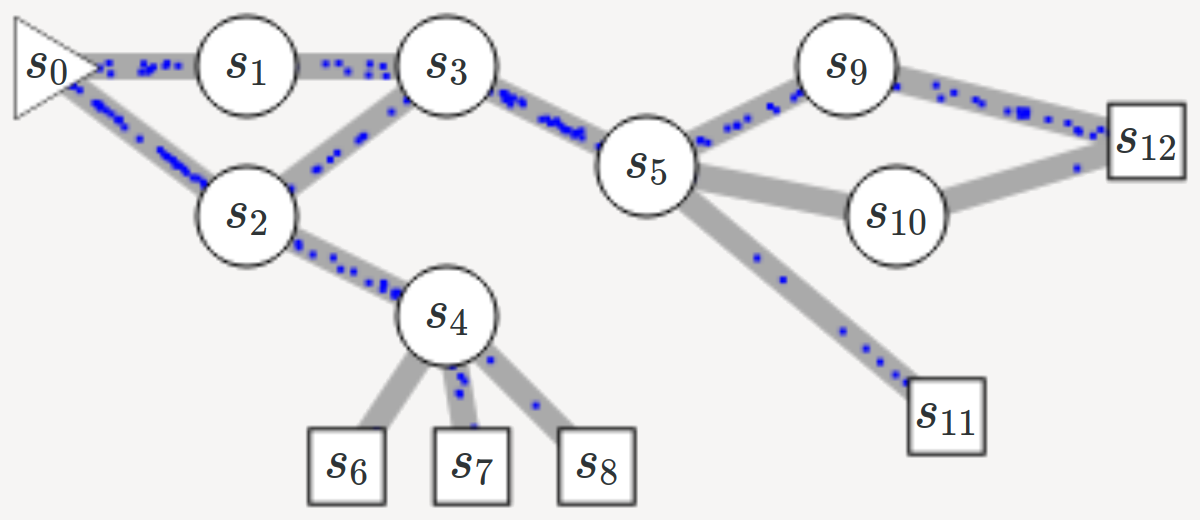] ] --- ## Methodology ### Algorithm .context[An **active learning** pipeline for DNA aptamer design] .right-column-66[.center[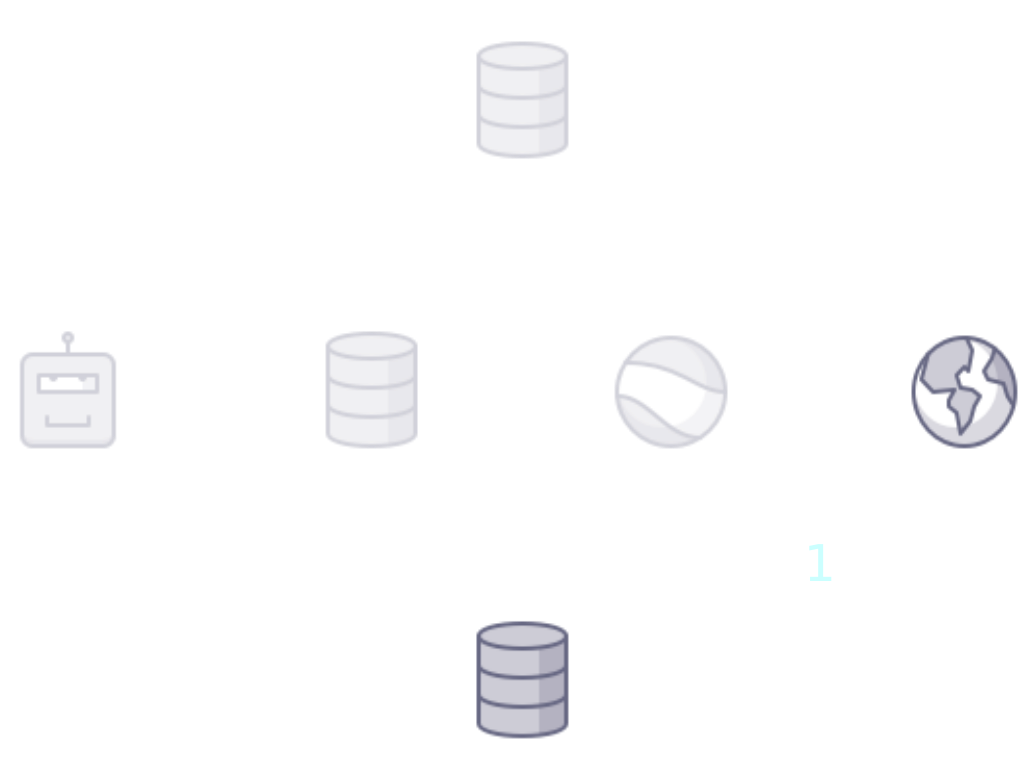]] .left-column-33[ **1**: Generate a _small_ initial _labelled_ data set from the oracle: $$\mathcal{D} = \mathcal{D_0}$$ In our main experiments: * $|\mathcal{D_0}| = 100$ * Length: 10-120 * Alphabet: 4 ] --- count: false ## Methodology ### Algorithm .context[An **active learning** pipeline for DNA aptamer design] .right-column-66[.center[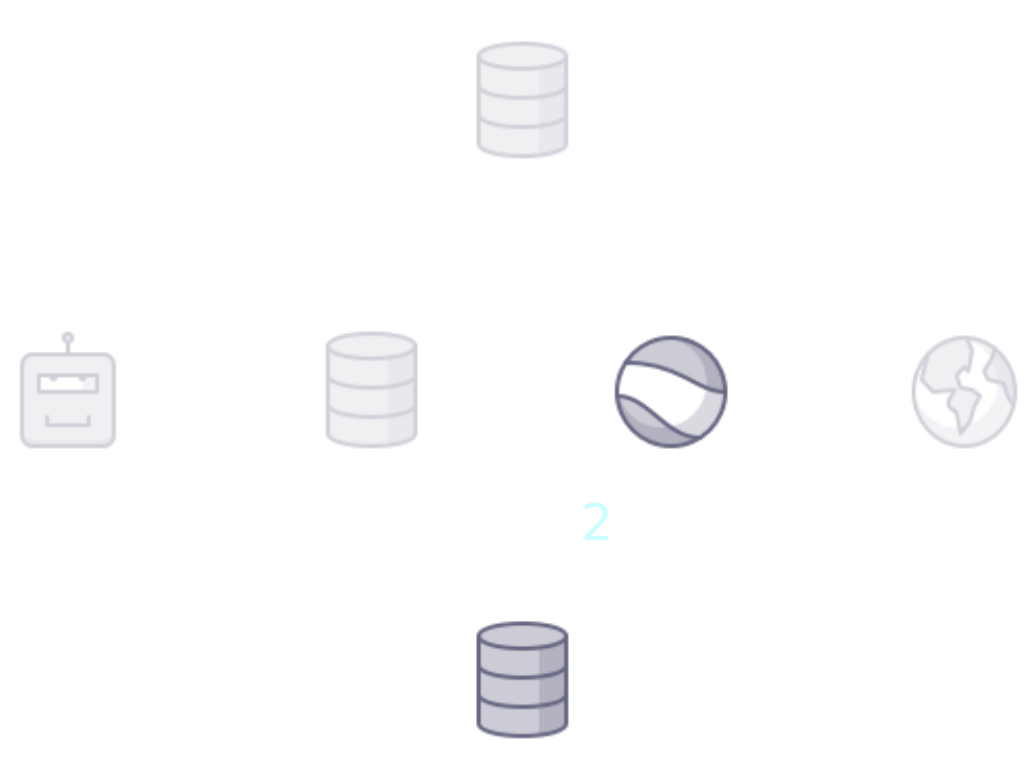]] .left-column-33[ **2**: Train model by minimising the error on $\mathcal{D}$: $$\min L(f_{\theta}(\mathcal{D}))$$ In our main experiments we train an ensemble of MLPs that provides with both _energy_ and _uncertainty_. ] --- count: false ## Methodology ### Algorithm .context[An **active learning** pipeline for DNA aptamer design] .right-column-66[.center[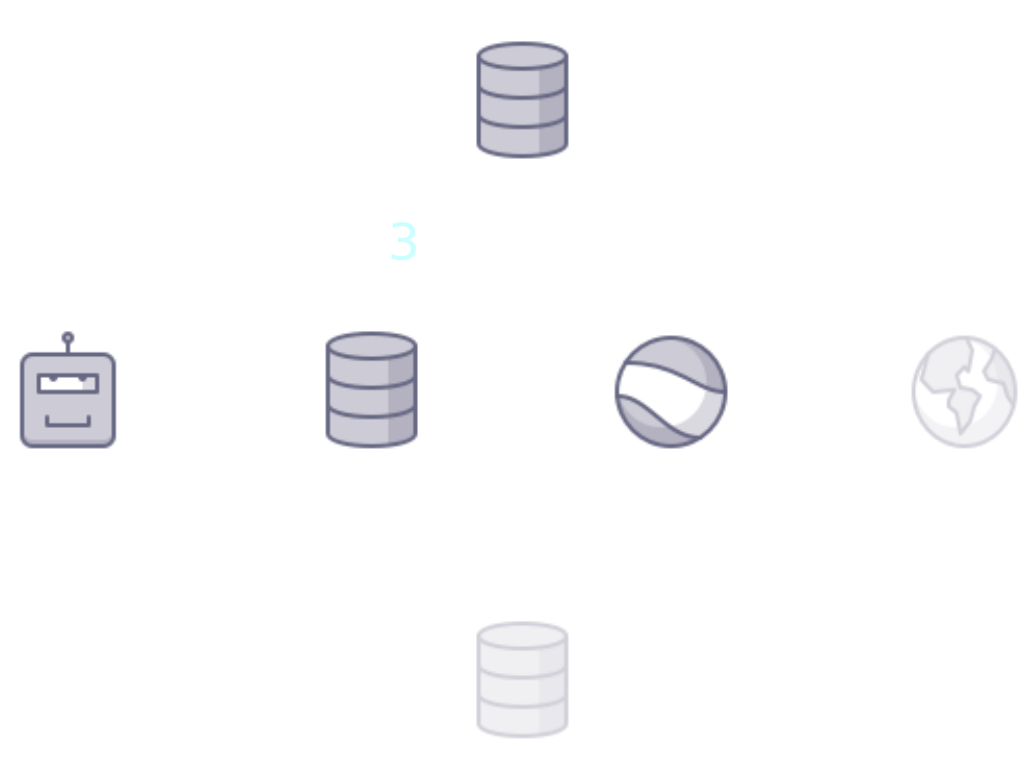]] .left-column-33[ **3**: Train GFlowNet until convergence using the ML model as a proxy oracle: .center[] .smaller[ $$ \sum^{s,a:T(s,a)=s'} Q(s,a) = R(s')+ \sum^{a' \in \mathcal{A}(s')} Q(s', a') $$ ] ] --- count: false ## Methodology ### Algorithm .context[An **active learning** pipeline for DNA aptamer design] .right-column-66[.center[]] .left-column-33[ **4**: Generate samples with GFlowNet and select a query of best candidates In our main experiments we select 100 samples per iteration and explore several selection methods (based on _energy_, _uncertainty_, both, a learnt policy, etc.) ] --- count: false ## Methodology ### Algorithm .context[An **active learning** pipeline for DNA aptamer design] .right-column-66[.center[]] .left-column-33[ **5**: Score the selected query with the oracle ($\mathcal{D_1}$) and add to the data set: $$\mathcal{D} = \mathcal{D} \cup \mathcal{D_1}$$ Then repeat all steps for $k$ iterations. ] --- ## Preliminary results .bigger[ * For simple oracles, the active learning pipeline generates the highest-reward samples in a few grand iterations. * For more complex oracles and longer aptamer sequences: * More iterations are requires * Query selection methods (annealing) become essential ] --- ## Current directions .bigger[ * Analyse the capabilities of the active learning pipeline and GFlowNet for hard to optimise functions. * Can GFlowNet be of help when MCMC is not enough? * What are the best methods for query selection? * Evaluate the pipeline with more sophisticated, real-world oracles .cite[([Kilgour et al., 2021](https://chemrxiv.org/engage/chemrxiv/article-details/60cbcb1d461f5627524764ab))] ] --- ## Application in other domains ### Electrocatalysts design .context[A similar active learning pipeline can be applied for **accelerating scientific discovery** in other domains.] .left-column-33[ <br><br> .center[] In collaboration with Marta Skreta, Victor Schmidt and David Rolnick. ] .right-column-66[.center[]] --- ## Call for postdoc applications If you are looking for a postdoc position and are interest in developing active learning models for chemical design, in a collaboration between McGill, Mila and NRC, contact Professor Lena Simine: .center[`lena.simine@mcgill.ca`] .center[  ] --- name: title class: title, middle ### Thanks! 감사합니다 .smaller[Michael Kilgour, Alex Hernández-García, Danny Salem, Tao Liu, Miroslava Cuperlovic-Culf, Yoshua Bengio, Lena Simine] .turquoise[Samsung-Mila-NYU Workshop · Online · Nov. 3rd 2021] .center[ <a href="https://mila.quebec/"><img src="../assets/images/slides/logos/mila-beige.png" alt="Mila" style="height: 4em"></a> <a href="http://www.umontreal.ca/"><img src="../assets/images/slides/logos/udem-white.png" alt="UdeM" style="height: 4em"></a> ] .footer[[alexhernandezgarcia.github.io](https://alexhernandezgarcia.github.io/) | [alex.hernandez-garcia@mila.quebec](mailto:alex.hernandez-garcia@mila.quebec) | [@alexhdezgcia](https://twitter.com/alexhdezgcia)] [](https://twitter.com/alexhdezgcia) --- .center[]