name: rolnicklab-apr24 class: title, middle ## Active learning for scientific discoveries Presenting: Alex Hernández-García (he/il/él) .smaller[Working with: David Rolnick, Nikita Saxena, Alex Duval, Victor Schmidt, Chritina Humer, Pierre Luc Carrier, Moksh Jain, Yoshua Bengio...] .turquoise[Rolnick's Lab, Mila · April 5th 2024] .center[ <a href="https://mila.quebec/"><img src="../assets/images/slides/logos/mila-beige.png" alt="Mila" style="width: 12em"></a> <a href="https://www.umontreal.ca/"><img src="../assets/images/slides/logos/udem-white.png" alt="UdeM" style="width: 12em"></a> <a href="https://www.umontreal.ca/"><img src="../assets/images/slides/logos/mcgill.png" alt="McGill" style="width: 12em"></a> ] .smaller[.footer[ Slides: [alexhernandezgarcia.github.io/slides/{{ name }}](https://alexhernandezgarcia.github.io/slides/{{ name }}) ]] --- ## Why machine learning for scientific discoveries? .left-column-66[ <br><br><br> - Mitigation of the climate crisis demands innovation in the energy and materials sector: .highlight1[materials discovery]. - The threat of antibiotic resistance and of new global pandemics demands innovation in .highlight1[drugs discovery]. - Other scientific disciplines can potentially benefit from the progress in scientific discovery pipelines. ] .right-column-33[ <br><br> .center[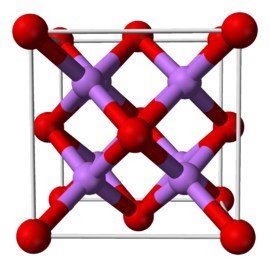] .center[] ] --- ## Traditional discovery cycle .context35[The climate crisis demands innovation in the scientific discovery model.] -- .right-column-66[<br>.center[]] .left-column-33[ <br> The .highlight1[traditional pipeline] for scientific discovery: * relies on .highlight1[highly specialised human expertise], * it is .highlight1[time-consuming] and * .highlight1[financially and computationally expensive]. ] --- count: false ## Machine learning in the loop .context35[The traditional scientific discovery loop is too slow for certain applications.] .right-column-66[<br>.center[]] .left-column-33[ <br> A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and ] --- count: false ## Machine learning in the loop .context35[The traditional scientific discovery loop is too slow for certain applications.] .right-column-66[<br>.center[]] .left-column-33[ <br> A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and * used to quickly and cheaply evaluate queries ] --- count: false ## Machine learning in the loop .context35[The traditional scientific discovery loop is too slow for certain applications.] .right-column-66[<br>.center[]] .left-column-33[ <br> A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and * used to quickly and cheaply evaluate queries .conclusion[There are infinitely many conceivable materials, $10^{180}$ potentially stable and $10^{60}$ drug molecules. Are predictive models enough?] ] --- count: false ## _Generative_ machine learning in the loop .right-column-66[<br>.center[]] .left-column-33[ <br> .highlight1[Generative machine learning] can: * .highlight1[learn structure] from the available data, * .highlight1[generalise] to unexplored regions of the search space and * .highlight1[build better queries] ] --- count: false ## _Generative_ machine learning in the loop .right-column-66[<br>.center[]] .left-column-33[ <br> .highlight1[Generative machine learning] can: * .highlight1[learn structure] from the available data, * .highlight1[generalise] to unexplored regions of the search space and * .highlight1[build better queries] .conclusion[Active learning with generative machine learning can in theory more efficiently explore the candidate space.] ] --- ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- ## Results - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. -- .center[] --- count: false ## Results - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Results - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Results - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Results - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Results - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Results - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- name: rolnicklab-apr24 class: title, middle  Alex Hernández-García (he/il/él) .center[ <a href="https://mila.quebec/"><img src="../assets/images/slides/logos/mila-beige.png" alt="Mila" style="height: 4em"></a> <a href="https://www.umontreal.ca/"><img src="../assets/images/slides/logos/udem-white.png" alt="UdeM" style="height: 4em"></a> ] .footer[[alexhernandezgarcia.github.io](https://alexhernandezgarcia.github.io/) | [alex.hernandez-garcia@mila.quebec](mailto:alex.hernandez-garcia@mila.quebec)]<br> .footer[[@alexhg@scholar.social](https://scholar.social/@alexhg) [](https://scholar.social/@alexhg) | [@alexhdezgcia](https://twitter.com/alexhdezgcia) [](https://twitter.com/alexhdezgcia)] .smaller[.footer[ Slides: [alexhernandezgarcia.github.io/slides/{{ name }}](https://alexhernandezgarcia.github.io/slides/{{ name }}) ]]