name: gflownets-jun24 class: title, middle ### GFlowNets tutorial Alex Hernández-García (he/il/él) .turquoise[Mila, Montreal · June 10th 2024] .center[ <a href="https://mila.quebec/"><img src="../assets/images/slides/logos/mila-beige.png" alt="Mila" style="height: 4em"></a> <a href="https://www.umontreal.ca/"><img src="../assets/images/slides/logos/udem-white.png" alt="UdeM" style="height: 4em"></a> ] .smaller[.footer[ Slides: [alexhernandezgarcia.github.io/slides/{{ name }}](https://alexhernandezgarcia.github.io/slides/{{ name }}) ]] .smaller[.smaller[.footer[ Note: This slide deck is largely [reused](https://alexhernandezgarcia.github.io/slides/futurehorizons-may24) and has more content than will be covered during this tutorial. ]]] --- count: false name: title class: title, middle ### Why scientific discoveries? .center[] --- ## Why scientific discoveries? .context[Climate change is a major challenge for humanity.] .center[ <figure> <img src="../assets/images/slides/climatechange/anthropogenic_temperature_rise.png" alt="Historical global average temperature and the influence of modern humans" style="width: 60%"> <figcaption>.smaller[Modelled and observed global average temperatures in the last 2 millenia (source graphic: <a href="https://www.theguardian.com/science/2021/aug/09/humans-have-caused-unprecedented-and-irreversible-change-to-climate-scientists-warn">The Guardian</a>.)]</figcaption> </figure> ] .conclusion["The evidence is clear: the time for action is now." .smaller[IPCC Report, 2022]] --- ## Why scientific discoveries? .context[Climate change is a major challenge for humanity.] .center[ <figure> <img src="../assets/images/slides/climatechange/who_climate_health.png" alt="Climate change presents a fundamental threat to human health." style="width: 100%"> <figcaption>.smaller[Climate-sensitive health risks (Adapted from: <a href="https://www.who.int/news-room/fact-sheets/detail/climate-change-and-health">World Health Organization</a>.)]</figcaption> </figure> ] .smaller[ * Environmental factors take the lives of around 13 million people _per year_. * Climate change affects people’s mental and physical health, access to clean air, safe water, food and health care. ] .full-width[ .conclusion["Climate change is the single biggest health threat facing humanity." .smaller[[WHO and WMO](https://climahealth.info/), 2024]] ] --- ## Why scientific discoveries? .center[] --- count: false ## Why scientific discoveries? .center[] -- .conclusion[Tackling climate change has a direct positive impact on global health. There is a strong synergy between machine learning research for climate and health.] --- ## Outline -- - [Machine learning for scientific discoveries to tackle climate and health challenges](#mlforscience) -- - [Gentle introduction to GFlowNets](#gflownets) -- - [Crystal-GFN: materials discovery](#crystal-gfn) -- - [Multi-fidelity active learning: drug and materials discovery](#mfal) --- count: false name: mlforscience class: title, middle ### Machine learning for scientific discoveries .center[] --- ## Traditional discovery cycle .context35[Climate and health challenges demand accelerating scientific discoveries.] -- .right-column-66[<br>.center[]] .left-column-33[ <br> The .highlight1[traditional pipeline] for scientific discovery: * relies on .highlight1[highly specialised human expertise], * it is .highlight1[time-consuming] and * .highlight1[financially and computationally expensive]. ] --- count: false ## _Active_ (machine) learning .context35[The traditional scientific discovery loop is too slow for certain applications.] .right-column-66[<br>.center[]] .left-column-33[ <br> A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and ] --- count: false ## _Active_ (machine) learning .context35[The traditional scientific discovery loop is too slow for certain applications.] .right-column-66[<br>.center[]] .left-column-33[ <br> A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and * used to quickly and cheaply evaluate queries ] --- count: false ## _Active_ (machine) learning .context35[The traditional scientific discovery loop is too slow for certain applications.] .right-column-66[<br>.center[]] .left-column-33[ <br> A .highlight1[machine learning model] can be: * trained with data from _real-world_ experiments and * used to quickly and cheaply evaluate queries .conclusion[There are infinitely many conceivable materials, $10^{180}$ potentially stable and $10^{60}$ drug molecules. Are predictive models enough?] ] --- count: false ## Active and _generative_ machine learning .right-column-66[<br>.center[]] .left-column-33[ <br> .highlight1[Generative machine learning] can: * .highlight1[learn structure] from the available data, * .highlight1[generalise] to unexplored regions of the search space and * .highlight1[build better queries] ] --- count: false ## Active and _generative_ machine learning .right-column-66[<br>.center[]] .left-column-33[ <br> .highlight1[Generative machine learning] can: * .highlight1[learn structure] from the available data, * .highlight1[generalise] to unexplored regions of the search space and * .highlight1[build better queries] .conclusion[Active learning with generative machine learning can in theory more efficiently explore the candidate space.] ] --- count: false name: title class: title, middle ### The challenges of scientific discoveries .center[] .center[] --- ## An intuitive trivial problem .highlight1[Problem]: find one arrangement of Tetris pieces on the board that minimise the empty space. .left-column-33[ .center[] ] .right-column-66[ .center[] ] -- .full-width[.center[ <figure> <img src="../assets/images/slides/tetris/mode1.png" alt="Score 12" style="width: 3%"> <figcaption>Score: 12</figcaption> </figure> ]] --- count: false ## An intuitive ~~trivial~~ easy problem .highlight1[Problem]: find .highlight2[all] the arrangements of Tetris pieces on the board that minimise the empty space. .left-column-33[ .center[] ] .right-column-66[ .center[] ] -- .full-width[.center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode1.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode2.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode3.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode4.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/mode5.png" alt="Score 12" style="width: 20%"> <figcaption>12</figcaption> </figure> </div> </div> ]] --- count: false ## An intuitive ~~easy~~ hard problem .highlight1[Problem]: find .highlight2[all] the arrangements of Tetris pieces on the board that minimise the empty space. .left-column-33[ .center[] ] .right-column-66[ .center[] ] -- .full-width[.center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode1.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode2.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode3.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode4.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/mode5.png" alt="Random board" style="width: 40%"> </figure> </div> </div> ]] --- count: false ## An incredibly ~~intuitive easy~~ hard problem .highlight1[Problem]: find .highlight2[all] the arrangements of Tetris pieces on the board that .highlight2[optimise an unknown function]. .left-column-33[ .center[] ] .right-column-66[ .center[] ] -- .full-width[.center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_434.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_800.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_815.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_849.png" alt="Random board" style="width: 40%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_905.png" alt="Random board" style="width: 40%"> </figure> </div> </div> ]] --- count: false ## An incredibly ~~intuitive easy~~ hard problem .highlight1[Problem]: find .highlight2[all] the arrangements of Tetris pieces on the board that .highlight2[optimise an unknown function]. .left-column-33[ .center[] ] .right-column-66[ .center[] ] .full-width[.conclusion[Materials and drug discovery involve finding candidates with rare properties from combinatorially or infinitely many options.]] --- ## Why Tetris for scientific discovery? .context35[The "Tetris problem" involves .highlight1[sampling from an unknown distribution] in a .highlight1[discrete, high-dimensional, combinatorially large space].] --- count: false ## Why Tetris for scientific discovery? ### Biological sequence design <br> Proteins, antimicrobial peptides (AMP) and DNA can be represented as sequences of amino acids or nucleobases. There are $22^{100} \approx 10^{134}$ protein sequences with 100 amino acids. .context35[The "Tetris problem" involves sampling from an unknown distribution in a discrete, high-dimensional, combinatorially large space] .center[] -- .left-column-66[ .dnag[`G`].dnaa[`A`].dnag[`G`].dnag[`G`].dnag[`G`].dnac[`C`].dnag[`G`].dnaa[`A`].dnac[`C`].dnag[`G`].dnag[`G`].dnat[`T`].dnaa[`A`].dnac[`C`].dnag[`G`].dnag[`G`].dnaa[`A`].dnag[`G`].dnac[`C`].dnat[`T`].dnac[`C`].dnat[`T`].dnag[`G`].dnac[`C`].dnat[`T`].dnac[`C`].dnac[`C`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`]<br> .dnat[`T`].dnac[`C`].dnaa[`A`].dnac[`C`].dnac[`C`].dnat[`T`].dnac[`C`].dnac[`C`].dnac[`C`].dnag[`G`].dnaa[`A`].dnag[`G`].dnac[`C`].dnaa[`A`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnag[`G`].dnat[`T`].dnat[`T`].dnag[`G`].dnat[`T`].dnaa[`A`].dnag[`G`].dnag[`G`].dnac[`C`].dnaa[`A`].dnag[`G`].dnac[`C`].dnag[`G`].dnat[`T`].dnac[`C`].dnac[`C`].dnat[`T`].dnaa[`A`].dnac[`C`].dnac[`C`].dnag[`G`].dnat[`T`].dnat[`T`].dnac[`C`].dnag[`G`]<br> .dnac[`C`].dnat[`T`].dnaa[`A`].dnac[`C`].dnag[`G`].dnac[`C`].dnag[`G`].dnat[`T`].dnac[`C`].dnat[`T`].dnac[`C`].dnat[`T`].dnat[`T`].dnat[`T`].dnac[`C`].dnag[`G`].dnag[`G`].dnag[`G`].dnag[`G`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`]<br> .dnat[`T`].dnat[`T`].dnag[`G`].dnac[`C`].dnaa[`A`].dnag[`G`].dnaa[`A`].dnag[`G`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`].dnaa[`A`].dnac[`C`].dnag[`G`].dnac[`C`].dnag[`G`].dnac[`C`].dnaa[`A`].dnat[`T`].dnag[`G`].dnac[`C`].dnag[`G`].dnaa[`A`].dnac[`C`].dnat[`T`].dnag[`G`].dnag[`G`].dnag[`G`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`].dnag[`G`].dnat[`T`].dnaa[`A`].dnag[`G`].dnat[`T`].dnac[`C`].dnag[`G`].dnaa[`A`].dnaa[`A`].dnac[`C`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnat[`T`].dnat[`T`].dnag[`G`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnaa[`A`].dnaa[`A`].dnac[`C`].dnaa[`A`]<br> .dnag[`G`].dnac[`C`].dnat[`T`].dnac[`C`].dnag[`G`].dnac[`C`].dnat[`T`].dnat[`T`].dnaa[`A`].dnag[`G`].dnag[`G`].dnag[`G`].dnac[`C`].dnac[`C`].dnat[`T`].dnac[`C`].dnag[`G`].dnaa[`A`].dnac[`C`].dnat[`T`].dnac[`C`].dnac[`C`].dnat[`T`].dnac[`C`].dnat[`T`].dnag[`G`].dnaa[`A`].dnaa[`A`].dnat[`T`].dnag[`G`].dnag[`G`].dnaa[`A`].dnag[`G`].dnat[`T`].dnag[`G`].dnat[`T`].dnat[`T`].dnac[`C`].dnaa[`A`].dnat[`T`].dnac[`C`].dnag[`G`].dnaa[`A`].dnaa[`A`].dnat[`T`].dnag[`G`].dnag[`G`].dnaa[`A`].dnag[`G`].dnat[`T`].dnag[`G`]<br> ] --- ## Why Tetris for scientific discovery? ### Molecular generation .context35[The "Tetris problem" involves sampling from an unknown distribution in a discrete, high-dimensional, combinatorially large space] <br> Small molecules can also be represented as sequences or by a combination of of higher-level fragments. There may be about $10^{60}$ drug-like molecules. -- .columns-3-left[ .center[  `CC(=O)NCCC1=CNc2c1cc(OC)cc2 CC(=O)NCCc1c[nH]c2ccc(OC)cc12` ]] .columns-3-center[ .center[  `OCCc1c(C)[n+](cs1)Cc2cnc(C)nc2N` ]] .columns-3-right[ .center[  `CN1CCC[C@H]1c2cccnc2` ]] --- ## Machine learning for scientific discovery ### Challenges and limitations of existing methods -- .highlight1[Challenge]: very large and high-dimensional search spaces. -- → Need for .highlight2[efficient search and generalisation] of underlying structure. -- .highlight1[Challenge]: underspecification of objective functions or metrics. -- → Need for .highlight2[diverse] candidates. -- .highlight1[Limitation]: Reinforcement learning excels at optimisation in complex spaces but tends to lack diversity. -- .highlight1[Limitation]: Markov chain Monte Carlo (MCMC) can _sample_ from a distribution (diversity) but struggles at mode mixing in high dimensions. -- → Need to .highlight2[combine all of the above]: sampling from complex, high-dimensional distributions. -- .conclusion[Generative flow networks (GFlowNets) address these challenges.] --- count: false name: gflownets class: title, middle ### A gentle introduction to GFlowNets .center[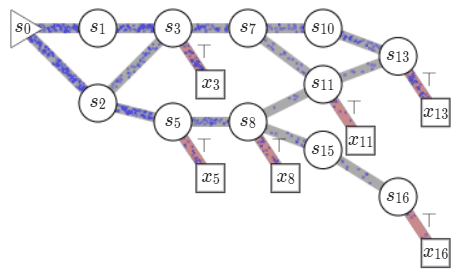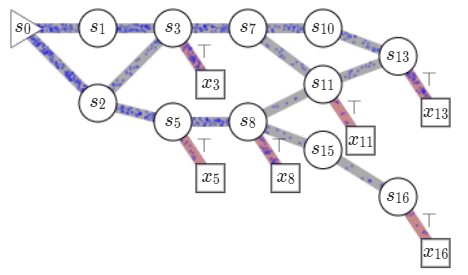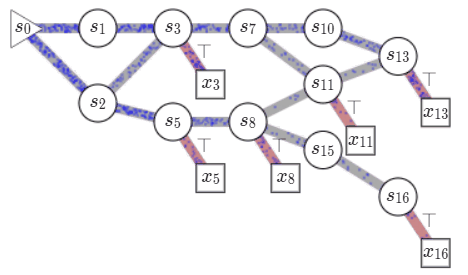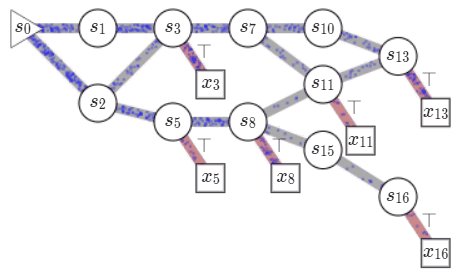] --- ## GFlowNets for science ### 3 key ingredients .context[Materials and drug discovery involve .highlight1[sampling from unknown distributions] in .highlight1[discrete or mixed, high-dimensional, combinatorially large spaces.]] -- <br><br> 1. .highlight1[Diversity] as an objective. -- - Given a score or reward function $R(x)$, learn to _sample proportionally to the reward_. -- 2. .highlight1[Compositionality] in the sample generation. -- - A meaningful decomposition of samples $x$ into multiple sub-states $s_0\rightarrow s_1 \rightarrow \dots \rightarrow x$ can yield generalisable patterns. -- 3. .highlight1[Deep learning] to learn from the generated samples. -- - A machine learning model can learn the transition function $F(s\rightarrow s')$ and generalise the patterns. --- ## 1. Diversity as an objective .context[Many existing approaches treat scientific discovery as an _optimisation_ problem.] <br> Given a reward or objective function $R(x)$, GFlowNet can be seen a generative model trained to sample objects $x \in \cal X$ according to .highlight1[a sampling policy $\pi(x)$ proportional to the reward $R(x)$]: -- .left-column[ $$\pi(x) = \frac{R(x)}{Z} \propto R(x)$$ ] -- .right-column[ $$Z = \sum_{x' \in \cal X} R(x')$$ ] -- .full-width[ .center[                                     ]] --- count: false ## 1. Diversity as an objective .context[Many existing approaches treat scientific discovery as an _optimisation_ problem.] <br> Given a reward or objective function $R(x)$, GFlowNet can be seen a generative model trained to sample objects $x \in \cal X$ according to .highlight1[a sampling policy $\pi(x)$ proportional to the reward $R(x)$]: .left-column[ $$\pi(x) = \frac{R(x)}{Z} \propto R(x)$$ ] .right-column[ $$Z = \sum_{x' \in \cal X} R(x')$$ ] .full-width[ → Sampling proportionally to the reward function enables finding .highlight1[multiple modes], hence .highlight1[diversity]. .center[] ] --- ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] The principle of compositionality is fundamental in semantics, linguistics, mathematical logic and is thought to be a cornerstone of human reasoning. --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. -- .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] .right-column[ <br><br> .conclusion[The decomposition of the sampling process into meaningful steps yields patterns that may be correlated with the reward function and facilitates learning complex distributions.] ] --- count: false ## 2. Compositionality ### Sample generation process .context35[Sampling _directly_ from a complex, high-dimensional distribution is difficult.] For the Tetris problem, a meaningful decomposition of the samples is .highlight1[adding one piece to the board at a time]. .left-column[.center[]] .right-column[ Objects $x \in \cal X$ are constructed through a sequence of actions from an .highlight1[action space $\cal A$]. ] .right-column[ At each step of the .highlight1[trajectory $\tau=(s_0\rightarrow s_1 \rightarrow \dots \rightarrow s_f)$], we get a partially constructed object $s$ in .highlight1[state space $\cal S$]. ] -- .right-column[ .conclusion[These ideas and terminology is reminiscent of reinforcement learning (RL).] ] --- ## 3. Deep learning policy .context35[GFlowNets learn a sampling policy $\pi\_{\theta}(x)$ proportional to the reward $R(x)$.] -- .left-column[ .center[] ] --- count: false ## 3. Deep learning policy .context35[GFlowNets learn a sampling policy $\pi\_{\theta}(x)$ proportional to the reward $R(x)$.] .left-column[ .center[] ] .right-column[ <br> Deep neural networks are trained to learn the transitions (flows) policy: $F\_{\theta}(s\_t\rightarrow s\_{t+1})$. ] -- .right-column[ Consistent flow theorem (informal): if the sum of the flows into state $s$ is equal to the sum of the flows out, then $\pi(x) \propto R(x)$. ] .references[ Bengio et al. [Flow network based generative models for non-iterative diverse candidate generation](https://arxiv.org/abs/2106.04399), NeurIPS, 2021. ] -- .right-column[ .conclusion[GFlowNets can be trained with deep learning methods to learn a sampling policy $\pi\_{\theta}$ proportional to a reward $R(x)$.] ] --- ## GFlowNets extensions and applications --- count: false ## GFlowNets extensions and applications ### Multi-objective GFlowNets Extension of GFlowNets to handle multi-objective optimisation and not only cover the Pareto front but also sample diverse objects at each point in the Pareto front. .center[] .references[ Jain et al. [Multi-Objective GFlowNets](https://arxiv.org/abs/2210.12765), ICML, 2023. ] --- ## GFlowNets extensions and applications ### Continuous GFlowNets Generalisation of the theory and implementation of GFlowNets to encompass both discrete and continuous or hybrid state spaces. .center[] .references[ Lahlou et al. [A Theory of Continuous Generative Flow Networks](https://arxiv.org/abs/2301.12594), ICML, 2023. ] --- ## GFlowNets extensions and applications ### Molecular conformation generation A continuous GFlowNets algorithm for sampling conformations of small molecules from the Boltzmann distribution, as determined by the molecule’s energy. .references[ Volokhova, Koziarski et al. [Towards equilibrium molecular conformation generation with GFlowNets](https://arxiv.org/abs/2310.14782), Digital Discovery, 2024. ] .center[] --- ## GFlowNets extensions and applications ### Biological sequence design An active learning algorithm with GFlowNets as a sampler of biological sequence design (DNA, antimicrobial peptides, proteins) with desirable properties. .center[] .left-column-66[ .dnag[`G`].dnaa[`A`].dnag[`G`].dnag[`G`].dnag[`G`].dnac[`C`].dnag[`G`].dnaa[`A`].dnac[`C`].dnag[`G`].dnag[`G`].dnat[`T`].dnaa[`A`].dnac[`C`].dnag[`G`].dnag[`G`].dnaa[`A`].dnag[`G`].dnac[`C`].dnat[`T`].dnac[`C`].dnat[`T`].dnag[`G`].dnac[`C`].dnat[`T`].dnac[`C`].dnac[`C`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`]<br> .dnat[`T`].dnac[`C`].dnaa[`A`].dnac[`C`].dnac[`C`].dnat[`T`].dnac[`C`].dnac[`C`].dnac[`C`].dnag[`G`].dnaa[`A`].dnag[`G`].dnac[`C`].dnaa[`A`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnag[`G`].dnat[`T`].dnat[`T`].dnag[`G`].dnat[`T`].dnaa[`A`].dnag[`G`].dnag[`G`].dnac[`C`].dnaa[`A`].dnag[`G`].dnac[`C`].dnag[`G`].dnat[`T`].dnac[`C`].dnac[`C`].dnat[`T`].dnaa[`A`].dnac[`C`].dnac[`C`].dnag[`G`].dnat[`T`].dnat[`T`].dnac[`C`].dnag[`G`]<br> .dnac[`C`].dnat[`T`].dnaa[`A`].dnac[`C`].dnag[`G`].dnac[`C`].dnag[`G`].dnat[`T`].dnac[`C`].dnat[`T`].dnac[`C`].dnat[`T`].dnat[`T`].dnat[`T`].dnac[`C`].dnag[`G`].dnag[`G`].dnag[`G`].dnag[`G`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`]<br> .dnat[`T`].dnat[`T`].dnag[`G`].dnac[`C`].dnaa[`A`].dnag[`G`].dnaa[`A`].dnag[`G`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`].dnaa[`A`].dnac[`C`].dnag[`G`].dnac[`C`].dnag[`G`].dnac[`C`].dnaa[`A`].dnat[`T`].dnag[`G`].dnac[`C`].dnag[`G`].dnaa[`A`].dnac[`C`].dnat[`T`].dnag[`G`].dnag[`G`].dnag[`G`].dnag[`G`].dnat[`T`].dnat[`T`].dnaa[`A`].dnag[`G`].dnat[`T`].dnaa[`A`].dnag[`G`].dnat[`T`].dnac[`C`].dnag[`G`].dnaa[`A`].dnaa[`A`].dnac[`C`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnat[`T`].dnat[`T`].dnag[`G`].dnaa[`A`].dnat[`T`].dnaa[`A`].dnaa[`A`].dnaa[`A`].dnac[`C`].dnaa[`A`]<br> .dnag[`G`].dnac[`C`].dnat[`T`].dnac[`C`].dnag[`G`].dnac[`C`].dnat[`T`].dnat[`T`].dnaa[`A`].dnag[`G`].dnag[`G`].dnag[`G`].dnac[`C`].dnac[`C`].dnat[`T`].dnac[`C`].dnag[`G`].dnaa[`A`].dnac[`C`].dnat[`T`].dnac[`C`].dnac[`C`].dnat[`T`].dnac[`C`].dnat[`T`].dnag[`G`].dnaa[`A`].dnaa[`A`].dnat[`T`].dnag[`G`].dnag[`G`].dnaa[`A`].dnag[`G`].dnat[`T`].dnag[`G`].dnat[`T`].dnat[`T`].dnac[`C`].dnaa[`A`].dnat[`T`].dnac[`C`].dnag[`G`].dnaa[`A`].dnaa[`A`].dnat[`T`].dnag[`G`].dnag[`G`].dnaa[`A`].dnag[`G`].dnat[`T`].dnag[`G`]<br> ] .references[ Jain et al. [Biological Sequence Design with GFlowNets](https://arxiv.org/abs/2203.04115), ICML, 2022. ] --- ## GFlowNets extensions and applications ### Review paper A review of the potential of GFlowNets for AI-driven scientific discoveries. .center[] .references[ Jain et al. [GFlowNets for AI-Driven Scientific Discovery](https://pubs.rsc.org/en/content/articlelanding/2023/dd/d3dd00002h). Digital Discovery, Royal Society of Chemistry, 2023. ] --- ## GFlowNet Python package Open sourced GFlowNet package, together with Mila collaborators: Nikita Saxena, Alexandra Volokhova, Michał Koziarski, Divya Sharma, Pierre Luc Carrier, Victor Schmidt, Joseph Viviano. .highlight2[Open source GFlowNet implementation]: [github.com/alexhernandezgarcia/gflownet](https://github.com/alexhernandezgarcia/gflownet) -- * A key design principle is the simplicity to create new environments. * Current environments: Tetris, hyper-grid, hyper-cube, hyper-torus, scrabble, crystals, molecules, DNA... * Discrete and continuous environments, multiple loss functions, etc. * Visualisation of results on WandDB --- count: false ## GFlowNet Python package Open sourced GFlowNet package, together with Mila collaborators: Nikita Saxena, Alexandra Volokhova, Michał Koziarski, Divya Sharma, Pierre Luc Carrier, Victor Schmidt, Joseph Viviano. .highlight2[Open source GFlowNet implementation]: [github.com/alexhernandezgarcia/gflownet](https://github.com/alexhernandezgarcia/gflownet) Research articles supported by this GFlowNet package: .smaller[ * Lahlou et al. [A Theory of Continuous Generative Flow Networks](https://arxiv.org/abs/2301.12594), ICML, 2023. * Hernandez-Garcia, Saxena et al. [Multi-fidelity active learning with GFlowNets](https://arxiv.org/abs/2306.11715). RealML, NeurIPS 2023. * Mila AI4Science et al. [Crystal-GFN: sampling crystals with desirable properties and constraints](https://arxiv.org/abs/2310.04925). AI4Mat, NeurIPS 2023 (spotlight). * Volokhova, Koziarski et al. [Towards equilibrium molecular conformation generation with GFlowNets](https://arxiv.org/abs/2310.14782). Digital Discovery, NeurIPS 2023. * Several other ongoing projects... ] --- count: false name: crystal-gfn class: title, middle ## Crystal-GFN: GFlowNets for materials discovery Mila AI4Science: Alex Hernandez-Garcia, Alexandre Duval, Alexandra Volokhova, Yoshua Bengio, Divya Sharma, Pierre Luc Carrier, Yasmine Benabed, Michał Koziarski, Victor Schmidt, Pierre-Paul De Breuck .smaller70[Mila AI4Science et al. [Crystal-GFN: sampling crystals with desirable properties and constraints](https://arxiv.org/abs/2310.04925). AI4Mat, NeurIPS 2023 (spotlight) / under review.] .center[] --- ## What are crystals? Definition: A crystal or crystalline solid is a solid material whose constituents (such as atoms, molecules, or ions) are arranged in a .highlight1[highly ordered microscopic structure], forming .highlight1[a crystal lattice that extends in all directions]. .left-column[ .center[] ] .right-column[ .center[] ] -- Here, we are concerned mainly with _inorganic crystals_, where the constituents are atoms or ions. -- A crystal structure is characterized by its .highlight1[unit cell], a small imaginary box containing atoms in a specific spatial arrangement with certain symmetry. The unit cell repeats iself periodically in all directions. --- ## Why do we care about crystals? .context35[Materials discovery can help reduce greenhouse gas emissions in multiple sectors.] -- <br> Many solid state materials are crystal structures and they are a core component of: * Electrocatalysts for fuel cells, hydrogen storage, industrial chemical reactions, carbon capture, etc. * Solid electrolytes for batteries. * Thin film materials for photovoltaics. * ... -- However, .highlight1[material modelling is very challenging]: * Limited data: only about 200 K known inorganic materials, but potentially $10^{180}$ possible stable materials (for reference: more than a billion molecules are known) * Sparsity: .highlight2[stable materials] only exist in a low-dimensional subspace of all possible 3D arrangements. -- .conclusion[There is a need for efficient generative models of crystal structures.] --- ## A domain-inspired approach ### Crystal structure parameters .context[Most previous works tackle crystal structure generation in the space of atomic coordinates and struggle to preserve the symmetry properties.] -- Instead of optimising the atom positions by learning from a small data set, we draw .highlight1[inspiration from theoretical crystallography to sample crystals in a lower-dimensional space of crystal structure parameters]. -- .highlight2[Space group]: symmetry operations of a repeating pattern in space that leave the pattern unchanged. -- - There are 17 symmetry groups in 2 dimensions (wallpaper groups). - There are 230 space groups in 3 dimensions. --- count: false ## A domain-inspired approach ### Crystal structure parameters .context[Most previous works tackle crystal structure generation in the space of atomic coordinates and struggle to preserve the symmetry properties.] Instead of optimising the atom positions by learning from a small data set, we draw .highlight1[inspiration from theoretical crystallography to sample crystals in a lower-dimensional space of crystal structure parameters]. .highlight2[Lattice system]: all 230 space groups can be classified into one of the 7 lattices. .center[ <div style="display: flex"> <div style="flex: 14%;"> <figure> <img src="../assets/images/slides/crystals/lattices/triclinic.png" alt="Triclinic" style="width: 100%"> <figcaption><small>Triclinic</small></figcaption> </figure> </div> <div style="flex: 14%;"> <figure> <img src="../assets/images/slides/crystals/lattices/monoclinic.png" alt="Monoclinic" style="width: 100%"> <figcaption><small>Monoclinic</small></figcaption> </figure> </div> <div style="flex: 14%;"> <figure> <img src="../assets/images/slides/crystals/lattices/orthorhombic.png" alt="Orthorhombic" style="width: 100%"> <figcaption><small>Orthorhombic</small></figcaption> </figure> </div> <div style="flex: 14%;"> <figure> <img src="../assets/images/slides/crystals/lattices/tetragonal.png" alt="Tetragonal" style="width: 100%"> <figcaption><small>Tetragonal</small></figcaption> </figure> </div> <div style="flex: 14%;"> <figure> <img src="../assets/images/slides/crystals/lattices/rhombohedral.png" alt="Rhombohedral" style="width: 100%"> <figcaption><small>Rhombohedral</small></figcaption> </figure> </div> <div style="flex: 14%;"> <figure> <img src="../assets/images/slides/crystals/lattices/hexagonal.png" alt="Hexagonal" style="width: 100%"> <figcaption><small>Hexagonal</small></figcaption> </figure> </div> <div style="flex: 14%;"> <figure> <img src="../assets/images/slides/crystals/lattices/cubic.png" alt="Cubic" style="width: 100%"> <figcaption><small>Cubic</small></figcaption> </figure> </div> </div> ] --- count: false ## A domain-inspired approach ### Crystal structure parameters .context[Most previous works tackle crystal structure generation in the space of atomic coordinates and struggle to preserve the symmetry properties.] Instead of optimising the atom positions by learning from a small data set, we draw .highlight1[inspiration from theoretical crystallography to sample crystals in a lower-dimensional space of crystal structure parameters]. .highlight2[Lattice parameters]: The lattice's size and shape is characterised by 6 parameters: .highlight1[$a, b, c, \alpha, \beta, \gamma$]. .center[] --- ## Crystal-GFlowNet ### Sequential generation .center[] --- count: false ## Crystal-GFlowNet ### Sequential generation .center[] --- count: false ## Crystal-GFlowNet ### Sequential generation .center[] --- count: false ## Crystal-GFlowNet ### Sequential generation .center[] --- count: false ## Crystal-GFlowNet ### Sequential generation .center[] --- count: false ## Crystal-GFlowNet ### Sequential generation .center[] --- count: false ## Crystal-GFlowNet ### Sequential generation .center[] --- count: false ## Crystal-GFlowNet ### Sequential generation .center[] --- count: false ## Crystal-GFlowNet ### Sequential generation .center[] --- count: false ## Crystal-GFlowNet ### Sequential generation .center[] .conclusion[Crystal-GFN binds multiple spaces representing crystallographic and material properties, setting intra- and inter-space hard constraints in the generation process.] --- ## GFlowNet approach ### Advantages .context[We generate materials in the lower-dimensional space of crystal structure parameters.] * Constructing materials by their crystal structure parameters allows us to introduce .highlight1[physicochemical and geometric _hard_ constraints]: -- * Charge neutrality of the composition. * Compatibility of composition and space group. * Hierarchical structure of the space group. * Compatibility of lattice parameters and lattice system. -- * .highlight1[Searching in the lower-dimensional space] of crystal structure parameters may be more efficient than in the space of atom coordinates. -- * Provided we have access to a predictive model of a material property, we can .highlight1[flexibly generate materials with desirable properties]. -- * We can .highlight1[flexibly sample materials with specific characteristics, such as composition or space group]. --- ## Crystal-GFlowNet ### Material properties We can train a Crystal-GFN with any reward function, provided it is computationally tractable. Therefore, we can use it to .highlight1[generate materials with different properties]. -- We have tested the following properties: - .highlight2[Formation energy] per atom [eV/atom], via a pre-trained machine learning model: indicative of the material's stability. -- - .highlight2[Electronic band gap] [eV] (squared distance to a target value, 1.34 eV), via a pre-trained machine learning model: relevant in photovoltaics, for instance. -- - Unit cell .highlight2[density] [g/cm<sup>3</sup>]: convenient as a proof of concept because we can calculate it _exactly_ from the GFN outputs. --- count: false ## Crystal-GFlowNet ### Material properties We can train a Crystal-GFN with any reward function, provided it is computationally tractable. Therefore, we can use it to .highlight1[generate materials with different properties]. We have tested the following properties: - .highlight2[Formation energy] per atom [eV/atom], via a pre-trained machine learning model: indicative of the material's stability. - .highlight2[Electronic band gap] [eV] (squared distance to a target value, 1.34 eV), via a pre-trained machine learning model: relevant in photovoltaics, for instance. - .alpha50[Unit cell .highlight2[density] [g/cm<sup>3</sup>]: convenient as a proof of concept because we can calculate it _exactly_ from the GFN outputs.] --- ## Results ### Formation energy .context35[The formation energy correlates with stability. The lower, the better.] .center[] --- count: false ## Results ### Formation energy .context35[The formation energy correlates with stability. The lower, the better.] .center[] --- count: false ## Results ### Formation energy .context35[The formation energy correlates with stability. The lower, the better.] .center[] --- count: false ## Results ### Formation energy .context35[The formation energy correlates with stability. The lower, the better.] .center[] --- count: false ## Results ### Formation energy .context[.highlight1[After training, Crystal-GFN samples structures with even lower formation energy [eV/atom] than the validation set.]] .center[] --- ## Results ### Band gap .context35[We aimed at sampling structures with band gap close to 1.34 eV.] .center[] --- count: false ## Results ### Band gap .context35[We aimed at sampling structures with band gap close to 1.34 eV.] .center[] --- count: false ## Results ### Band gap .context35[We aimed at sampling structures with band gap close to 1.34 eV.] .center[] --- count: false ## Results ### Band gap .context35[We aimed at sampling structures with band gap close to 1.34 eV.] .center[] --- count: false ## Results ### Band gap .context[.highlight1[After training, Crystal-GFN samples structures with band gap [eV] around the target value.]] .center[] --- ## Results ### Diversity .context[.highlight2[Diversity] is key in materials discovery.] Analysis of 10,000 sampled crystals and the top-100 with lowest formation energy. -- - All 10,000 samples are unique. -- - All crystal systems, lattice systems and point symmetries found in the 10,000 samples. - 4 out of 8 crystal-lattice systems in the top-100. - 4 out of the 5 point symmetries in the top-100. -- - All 22 elements found in the 10,000 samples. - 15 out of 22 elements in the top-100. -- - 73 out of 113 space groups (65 %) found in the 10,000 samples - 19 out of 113 space groups in the top-100. -- .conclusion[Crystal-GFN samples are highly diverse.] --- ## Crystal-GFN ### Summary and conclusions .references[ * Mila AI4Science et al. [Crystal-GFN: sampling crystals with desirable properties and constraints](https://arxiv.org/abs/2310.04925). AI4Mat, NeurIPS 2023 (spotlight). ] * Discovering new crystal structures with desirable properties can help mitigate the climate crisis. -- * There are infinitely many conceivable crystals. Only a few are stable. Only a few stable crystals have interesting properties. This is a really hard problem. -- * Most methods in the literature struggle to preserve the symmetry properties of the crystals. -- * Crystal-GFN introduces .highlight1[physicochemical and structural constraints], reducing the search space. * Crystal-GFN was trained in 30 hours in a CPU-only machine. -- * Our results show that we can generate .highlight1[diverse, high scoring samples with the desired constraints]. -- * The .highlight1[framework can be flexibly extended] with more constraints, crystal structure descriptors (atomic positions) and other properties. --- count: false name: mfal class: title, middle ## Multi-fidelity active learning Nikita Saxena, Moksh Jain, Cheng-Hao Liu, Yoshua Bengio .smaller[[Multi-fidelity active learning with GFlowNets](https://arxiv.org/abs/2306.11715). RealML, NeurIPS 2023 / under review.] .center[] --- ## Why multi-fidelity? .context35[We had described the scientific discovery loop as a cycle with one single oracle.] <br><br> .right-column[ .center[] ] -- .left-column[ Example: "incredibly hard" Tetris problem: find arrangements of Tetris pieces that optimise an .highlight2[unknown function $f$]. - $f$: Oracle, cost per evaluation 1000 CAD. .center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_434.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_800.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_815.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_849.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_905.png" alt="Random board" style="width: 200%"> </figure> </div> </div> ] ] --- count: false ## Why multi-fidelity? .context35[However, in practice, multiple oracles (models) of different fidelity and cost are available in scientific applications.] <br><br> .right-column[ .center[] ] .left-column[ Example: "incredibly hard" Tetris problem: find arrangements of Tetris pieces that optimise an .highlight2[unknown function $f$]. - $f$: Oracle, cost per evaluation 1000 CAD. .center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_434.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_800.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_815.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_849.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_905.png" alt="Random board" style="width: 200%"> </figure> </div> </div> ] ] --- count: false ## Why multi-fidelity? .context35[However, in practice, multiple oracles (models) of different fidelity and cost are available in scientific applications.] <br><br> .right-column[ .center[] ] .left-column[ Example: "incredibly hard" Tetris problem: find arrangements of Tetris pieces that optimise an .highlight2[unknown function $f$]. - $f$: Oracle, cost per evaluation 1000 CAD. - $f\_1$: Slightly inaccurate oracle, cost 100 CAD. - $f\_2$: Noisy but informative oracle, cost 1 CAD. .center[ <div style="display: flex"> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_434.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_800.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_815.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_849.png" alt="Random board" style="width: 200%"> </figure> </div> <div style="flex: 20%;"> <figure> <img src="../assets/images/slides/tetris/10x20/random_905.png" alt="Random board" style="width: 200%"> </figure> </div> </div> ] ] --- count: false ## Why multi-fidelity? .context[In many scientific applications we have access to multiple approximations of the objective function.] .left-column[ For example, in .highlight1[material discovery]: * .highlight1[Synthesis] of a material and characterisation of a property in the lab * Quantum mechanic .highlight1[simulations] to estimate the property * .highlight1[Machine learning] models trained to predict the property ] .right-column[ .center[] ] -- .conclusion[However, current machine learning methods cannot efficiently leverage the availability of multiple oracles and multi-fidelity data. Especially with .highlight1[structured, large, high-dimensional search spaces].] --- ## Contribution - An .highlight1[active learning] algorithm to leverage the availability of .highlight1[multiple oracles at different fidelities and costs]. -- - The goal is two-fold: 1. Find high-scoring candidates 2. Candidates must be diverse -- - Experimental evaluation with .highlight1[biological sequences and molecules]: - DNA - Antimicrobial peptides - Small molecules - Classical multi-fidelity toy functions (Branin and Hartmann) -- .conclusion[Likely the first multi-fidelity active learning method for biological sequences and molecules.] --- ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[] --- count: false ## Our multi-fidelity active learning algorithm .center[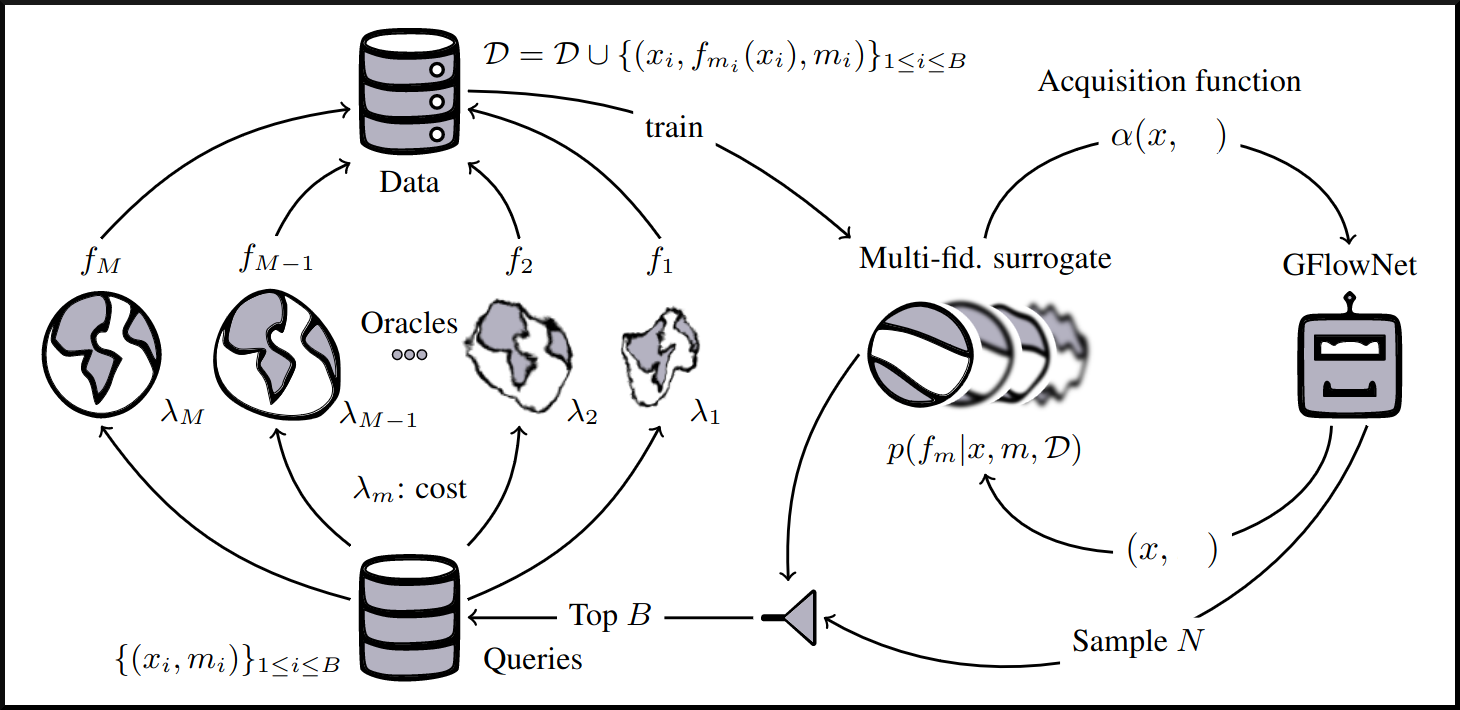] --- count: false ## Our multi-fidelity active learning algorithm .center[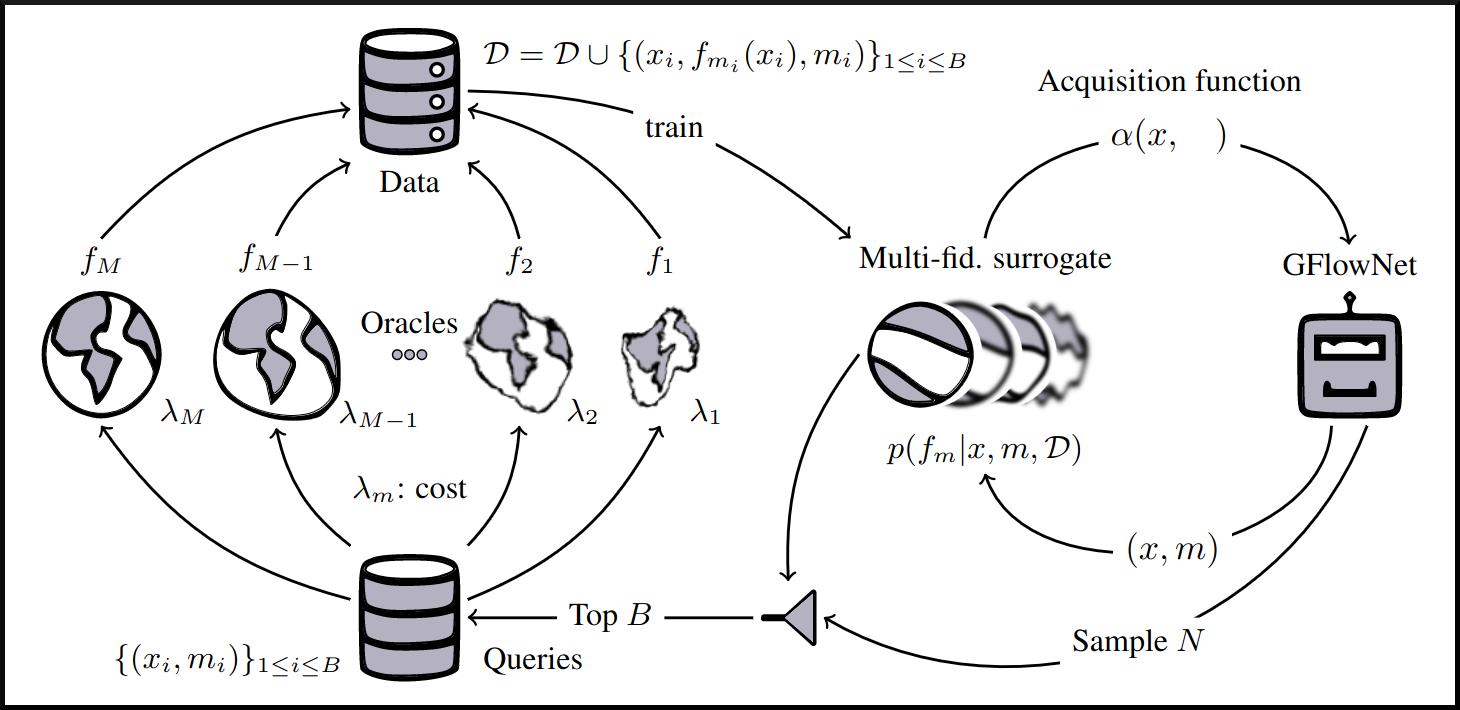] --- count: false ## Our multi-fidelity active learning algorithm .center[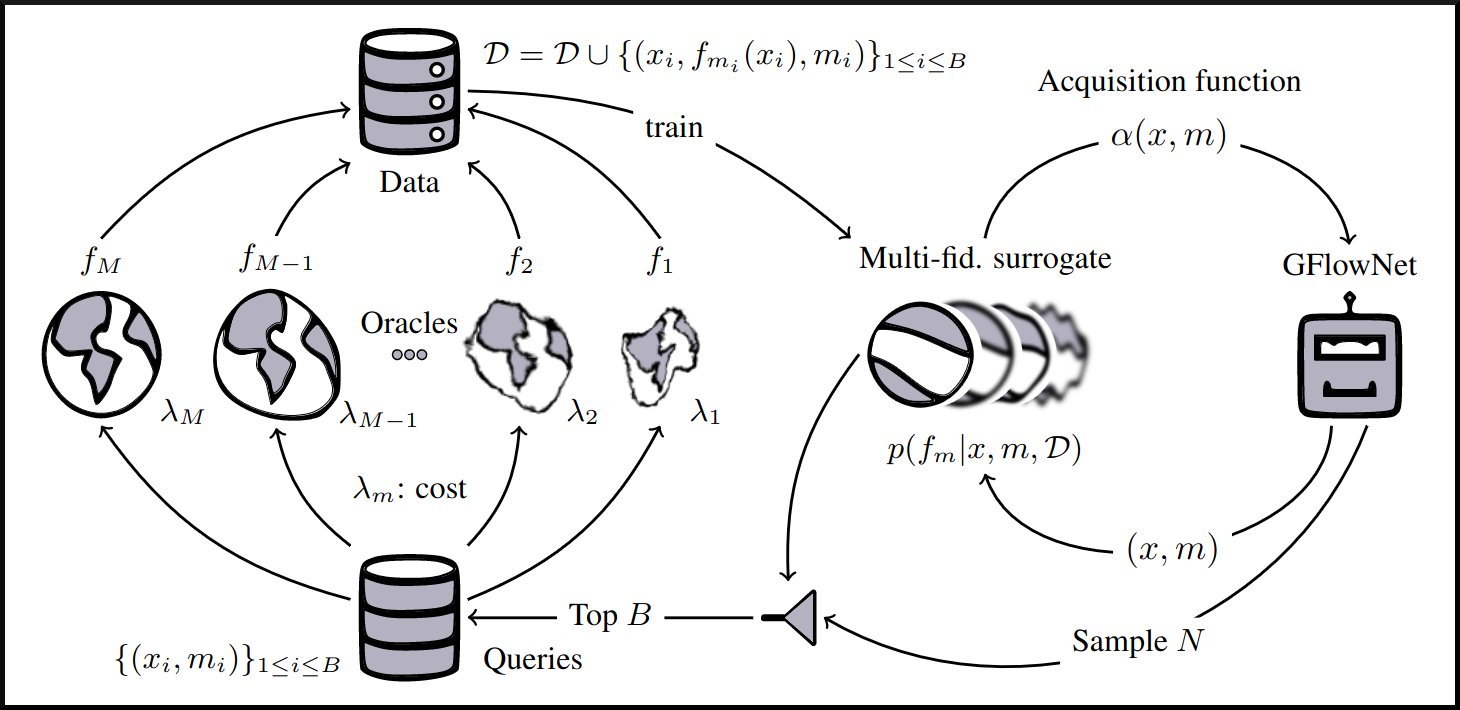] --- ## Experiments ### Baselines .context[This is the .highlight1[first multi-fidelity active learning algorithm tested on biological sequence design and molecular design problems]. There did not exist baselines from the literature.] -- <br> * .highlight1[SF-GFN]: GFlowNet with highest fidelity oracle to establish a benchmark for performance without considering the cost-accuracy trade-offs. -- * .highlight1[Random]: Quasi-random approach where the candidates and fidelities are picked randomly and the top $(x, m)$ pairs scored by the acquisition function are queried. -- * .highlight1[Random fid. GFN]: GFlowNet with random fidelities, to investigate the benefit of deciding the fidelity with GFlowNets. -- * .highlight1[MF-PPO]: Replacement of MF-GFN with a reinforcement learning algorithm to _optimise_ the acquisition function. --- ## Small molecules - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. -- .center[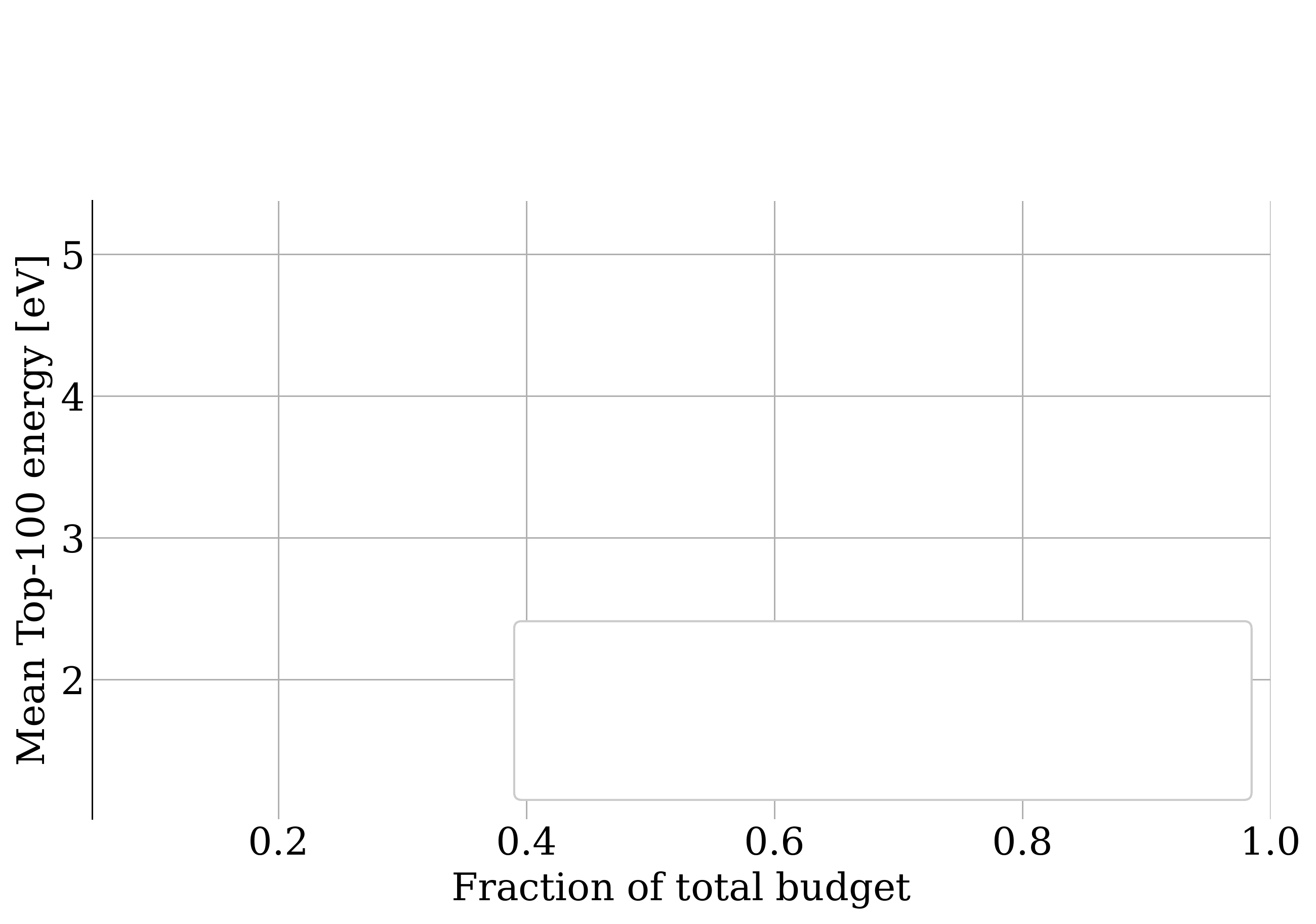] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic electron affinity (EA). Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- count: false ## Small molecules - Realistic experiments with experimental oracles and costs that reflect the computational demands (1, 3, 7). - GFlowNet adds one SELFIES token (out of 26) at a time with variable length up to 64 ($|\mathcal{X}| > 26^{64}$). - Property: Adiabatic .highlight1[ionisation potential (IP)]. Relevant in organic semiconductors, photoredox catalysis and organometallic synthesis. .center[] --- ## DNA aptamers - GFlowNet adds one nucleobase (`A`, `T`, `C`, `G`) at a time up to length 30. This yields a design space of size $|\mathcal{X}| = 4^{30}$. - The objective function is the free energy estimated by a bioinformatics tool. - The (simulated) lower fidelity oracle is a transformer trained with 1 million sequences. -- .center[] --- count: false ## Antimicrobial peptides (AMP) - Protein sequences (20 amino acids) with variable length (max. 50). - The oracles are 3 ML models trained with different subsets of data. -- .center[] --- ## How does multi-fidelity help? .context[Visualisation on the synthetic 2D Branin function task.] .center[] --- count: false ## How does multi-fidelity help? .context[Visualisation on the synthetic 2D Branin function task.] .center[] --- count: false ## How does multi-fidelity help? .context[Visualisation on the synthetic 2D Branin function task.] .center[] --- count: false ## How does multi-fidelity help? .context[Visualisation on the synthetic 2D Branin function task.] .center[] --- ## Multi-fidelity active learning with GFlowNets ### Summary and conclusions .references[ * Hernandez-Garcia, Saxena et al. [Multi-fidelity active learning with GFlowNets](https://arxiv.org/abs/2306.11715). RealML, NeurIPS 2023. ] * Current ML for science methods do not utilise all the information and resources at our disposal. -- * AI-driven scientific discovery demands learning methods that can .highlight1[efficiently discover diverse candidates in combinatorially large, high-dimensional search spaces]. -- * .highlight1[Multi-fidelity active learning with GFlowNets] enables .highlight1[cost-effective exploration] of large, high-dimensional and structured spaces, and discovers multiple, diverse modes of black-box score functions. -- * This is to our knowledge the first algorithm capable of effectively leveraging multi-fidelity oracles to discover diverse biological sequences and molecules. --- ## Acknowledgements .columns-3-left[ Victor Schmidt<br> Mélisande Teng<br> Alexandre Duval<br> Yasmine Benabed<br> Pierre Luc Carrier<br> Divya Sharma<br> Yoshua Bengio<br> Lena Simine<br> Michael Kilgour<br> ... ] .columns-3-center[ Alexandra Volokhova<br> Michał Koziarski<br> Paula Harder<br> David Rolnick<br> Qidong Yang<br> Santiago Miret<br> Sasha Luccioni<br> Alexia Reynaud<br> Tianyu Zhang<br> ... ] .columns-3-right[ Nikita Saxena<br> Moksh Jain<br> Cheng-Hao Liu<br> Kolya Malkin<br> Tristan Deleu<br> Salem Lahlou<br> Alvaro Carbonero<br> José González-Abad<br> Emmanuel Bengio<br> ... ] .conclusion[Science is a lot more fun when shared with bright and interesting people!] --- count: false name: title class: title, middle ## Overall summary and conclusions .center[] --- ## Summary and conclusions - Scientific discoveries can help us tackle the climate crisis and health challenges. -- - Machine learning has great potential to accelerate scientific discoveries. There are strong synergies between materials discovery and drug discovery methods. -- - With GFlowNets, we are able to address some important challenges: discover diverse candidates in very large, complex search spaces. -- - Crystal-GFN rethinks crystal structure generation by introducing domain knowledge and hard constraints to discover materials with desirable properties. -- - Multi-fidelity active learning with GFlowNets effectively leverages the availability of multiple oracles for the first time for certain scientific discovery problems. --- name: gflownets-jun24 class: title, middle  Alex Hernández-García (he/il/él) .center[ <a href="https://mila.quebec/"><img src="../assets/images/slides/logos/mila-beige.png" alt="Mila" style="height: 4em"></a> <a href="https://www.umontreal.ca/"><img src="../assets/images/slides/logos/udem-white.png" alt="UdeM" style="height: 4em"></a> ] .footer[[alexhernandezgarcia.github.io](https://alexhernandezgarcia.github.io/) | [alex.hernandez-garcia@mila.quebec](mailto:alex.hernandez-garcia@mila.quebec)]<br> .footer[[@alexhg@scholar.social](https://scholar.social/@alexhg) [](https://scholar.social/@alexhg) | [@alexhdezgcia](https://twitter.com/alexhdezgcia) [](https://twitter.com/alexhdezgcia)] .smaller[.footer[ Slides: [alexhernandezgarcia.github.io/slides/{{ name }}](https://alexhernandezgarcia.github.io/slides/{{ name }}) ]]